
Rhizorhabdus dicambivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Rhizorhabdus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
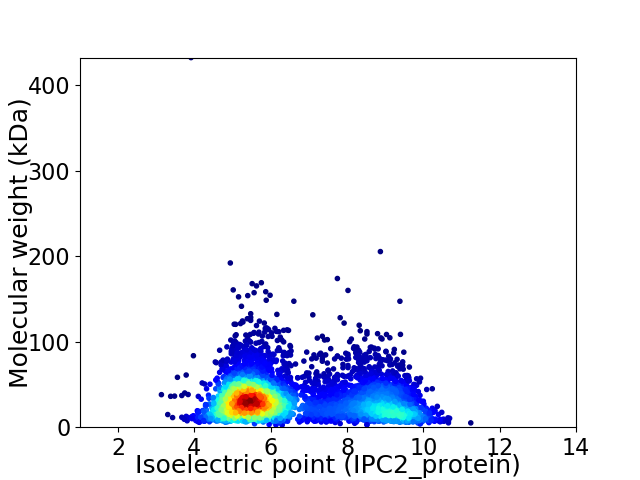
Virtual 2D-PAGE plot for 4856 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A4FZA5|A0A2A4FZA5_9SPHN TonB-dependent receptor OS=Rhizorhabdus dicambivorans OX=1850238 GN=COO09_07955 PE=3 SV=1
MM1 pKa = 7.19STPTSNSVTAEE12 pKa = 4.28RR13 pKa = 11.84ITLPADD19 pKa = 3.88PDD21 pKa = 4.02LSSLTHH27 pKa = 5.92GSRR30 pKa = 11.84WVATGATTSLTYY42 pKa = 10.58SFPINAGNWANPYY55 pKa = 10.45SPLNEE60 pKa = 4.17PSTGGVIPLDD70 pKa = 3.43AAAATGAVTALQLWSRR86 pKa = 11.84YY87 pKa = 10.03ANISFAATSDD97 pKa = 3.07TGANVGDD104 pKa = 3.33IRR106 pKa = 11.84FAYY109 pKa = 9.08TRR111 pKa = 11.84DD112 pKa = 3.19EE113 pKa = 4.3DD114 pKa = 4.3AAAHH118 pKa = 6.76AYY120 pKa = 10.15LPSSGPAAGDD130 pKa = 2.75IWLDD134 pKa = 3.36STTDD138 pKa = 3.55FGGFEE143 pKa = 4.08RR144 pKa = 11.84GSFGLFTLLHH154 pKa = 5.89EE155 pKa = 4.85VGHH158 pKa = 6.45ALGLKK163 pKa = 10.15HH164 pKa = 6.45PFEE167 pKa = 5.32SSTLNEE173 pKa = 4.17QILDD177 pKa = 3.89PEE179 pKa = 4.67LDD181 pKa = 4.0SVNSTVMSYY190 pKa = 11.21NLYY193 pKa = 10.65SDD195 pKa = 5.69LPASDD200 pKa = 4.44YY201 pKa = 11.23GISYY205 pKa = 10.56LPTTPMQLDD214 pKa = 3.88IEE216 pKa = 4.54AVQALYY222 pKa = 10.47GARR225 pKa = 11.84PFNTDD230 pKa = 2.89DD231 pKa = 3.46NEE233 pKa = 4.42YY234 pKa = 10.38VFNSGSQYY242 pKa = 10.75FEE244 pKa = 4.52TIYY247 pKa = 11.11DD248 pKa = 3.51SGGNDD253 pKa = 3.4TIIWNSSTEE262 pKa = 3.85EE263 pKa = 3.75AVIDD267 pKa = 4.33LEE269 pKa = 4.46SGSWSALGDD278 pKa = 3.66PLITFDD284 pKa = 4.18RR285 pKa = 11.84QGEE288 pKa = 4.6VVKK291 pKa = 10.89VDD293 pKa = 3.68YY294 pKa = 8.52FTVAIFSEE302 pKa = 4.37TMIEE306 pKa = 4.04NATGGGGDD314 pKa = 3.85DD315 pKa = 5.31LIYY318 pKa = 11.14GNAVANRR325 pKa = 11.84LLGSDD330 pKa = 3.21GHH332 pKa = 7.84DD333 pKa = 4.56DD334 pKa = 3.19IFGYY338 pKa = 10.57EE339 pKa = 4.16GADD342 pKa = 3.49TLVGGAGNDD351 pKa = 3.54HH352 pKa = 7.08LYY354 pKa = 10.95GRR356 pKa = 11.84AASGGADD363 pKa = 3.77GEE365 pKa = 4.72DD366 pKa = 4.05SLSGGAGSDD375 pKa = 3.62YY376 pKa = 11.17LQGNAGNDD384 pKa = 3.44MLDD387 pKa = 3.51GGEE390 pKa = 4.11GSDD393 pKa = 4.35RR394 pKa = 11.84INGGGSDD401 pKa = 3.74DD402 pKa = 5.18SIFGGAGNDD411 pKa = 3.61TVNGNLGSDD420 pKa = 4.16VISGGDD426 pKa = 3.35GDD428 pKa = 4.34DD429 pKa = 3.89SLRR432 pKa = 11.84GGQGADD438 pKa = 3.71SISGGTGNDD447 pKa = 3.2ILSGDD452 pKa = 4.03LGEE455 pKa = 6.02DD456 pKa = 3.1ILSGGDD462 pKa = 3.52GNDD465 pKa = 3.26LFVVGGSGSPLAQPDD480 pKa = 4.16RR481 pKa = 11.84ILDD484 pKa = 3.87FTDD487 pKa = 3.48GTDD490 pKa = 2.86WVAIGFAPAAILTGAAQANAGAAATTAQQLLDD522 pKa = 3.54NRR524 pKa = 11.84EE525 pKa = 4.17GNGEE529 pKa = 4.01VAALAVGADD538 pKa = 3.34TYY540 pKa = 11.59LFYY543 pKa = 11.08ASNGGGMIDD552 pKa = 3.41SAIVVVGIAPQAFGVADD569 pKa = 4.7FGG571 pKa = 4.47
MM1 pKa = 7.19STPTSNSVTAEE12 pKa = 4.28RR13 pKa = 11.84ITLPADD19 pKa = 3.88PDD21 pKa = 4.02LSSLTHH27 pKa = 5.92GSRR30 pKa = 11.84WVATGATTSLTYY42 pKa = 10.58SFPINAGNWANPYY55 pKa = 10.45SPLNEE60 pKa = 4.17PSTGGVIPLDD70 pKa = 3.43AAAATGAVTALQLWSRR86 pKa = 11.84YY87 pKa = 10.03ANISFAATSDD97 pKa = 3.07TGANVGDD104 pKa = 3.33IRR106 pKa = 11.84FAYY109 pKa = 9.08TRR111 pKa = 11.84DD112 pKa = 3.19EE113 pKa = 4.3DD114 pKa = 4.3AAAHH118 pKa = 6.76AYY120 pKa = 10.15LPSSGPAAGDD130 pKa = 2.75IWLDD134 pKa = 3.36STTDD138 pKa = 3.55FGGFEE143 pKa = 4.08RR144 pKa = 11.84GSFGLFTLLHH154 pKa = 5.89EE155 pKa = 4.85VGHH158 pKa = 6.45ALGLKK163 pKa = 10.15HH164 pKa = 6.45PFEE167 pKa = 5.32SSTLNEE173 pKa = 4.17QILDD177 pKa = 3.89PEE179 pKa = 4.67LDD181 pKa = 4.0SVNSTVMSYY190 pKa = 11.21NLYY193 pKa = 10.65SDD195 pKa = 5.69LPASDD200 pKa = 4.44YY201 pKa = 11.23GISYY205 pKa = 10.56LPTTPMQLDD214 pKa = 3.88IEE216 pKa = 4.54AVQALYY222 pKa = 10.47GARR225 pKa = 11.84PFNTDD230 pKa = 2.89DD231 pKa = 3.46NEE233 pKa = 4.42YY234 pKa = 10.38VFNSGSQYY242 pKa = 10.75FEE244 pKa = 4.52TIYY247 pKa = 11.11DD248 pKa = 3.51SGGNDD253 pKa = 3.4TIIWNSSTEE262 pKa = 3.85EE263 pKa = 3.75AVIDD267 pKa = 4.33LEE269 pKa = 4.46SGSWSALGDD278 pKa = 3.66PLITFDD284 pKa = 4.18RR285 pKa = 11.84QGEE288 pKa = 4.6VVKK291 pKa = 10.89VDD293 pKa = 3.68YY294 pKa = 8.52FTVAIFSEE302 pKa = 4.37TMIEE306 pKa = 4.04NATGGGGDD314 pKa = 3.85DD315 pKa = 5.31LIYY318 pKa = 11.14GNAVANRR325 pKa = 11.84LLGSDD330 pKa = 3.21GHH332 pKa = 7.84DD333 pKa = 4.56DD334 pKa = 3.19IFGYY338 pKa = 10.57EE339 pKa = 4.16GADD342 pKa = 3.49TLVGGAGNDD351 pKa = 3.54HH352 pKa = 7.08LYY354 pKa = 10.95GRR356 pKa = 11.84AASGGADD363 pKa = 3.77GEE365 pKa = 4.72DD366 pKa = 4.05SLSGGAGSDD375 pKa = 3.62YY376 pKa = 11.17LQGNAGNDD384 pKa = 3.44MLDD387 pKa = 3.51GGEE390 pKa = 4.11GSDD393 pKa = 4.35RR394 pKa = 11.84INGGGSDD401 pKa = 3.74DD402 pKa = 5.18SIFGGAGNDD411 pKa = 3.61TVNGNLGSDD420 pKa = 4.16VISGGDD426 pKa = 3.35GDD428 pKa = 4.34DD429 pKa = 3.89SLRR432 pKa = 11.84GGQGADD438 pKa = 3.71SISGGTGNDD447 pKa = 3.2ILSGDD452 pKa = 4.03LGEE455 pKa = 6.02DD456 pKa = 3.1ILSGGDD462 pKa = 3.52GNDD465 pKa = 3.26LFVVGGSGSPLAQPDD480 pKa = 4.16RR481 pKa = 11.84ILDD484 pKa = 3.87FTDD487 pKa = 3.48GTDD490 pKa = 2.86WVAIGFAPAAILTGAAQANAGAAATTAQQLLDD522 pKa = 3.54NRR524 pKa = 11.84EE525 pKa = 4.17GNGEE529 pKa = 4.01VAALAVGADD538 pKa = 3.34TYY540 pKa = 11.59LFYY543 pKa = 11.08ASNGGGMIDD552 pKa = 3.41SAIVVVGIAPQAFGVADD569 pKa = 4.7FGG571 pKa = 4.47
Molecular weight: 58.36 kDa
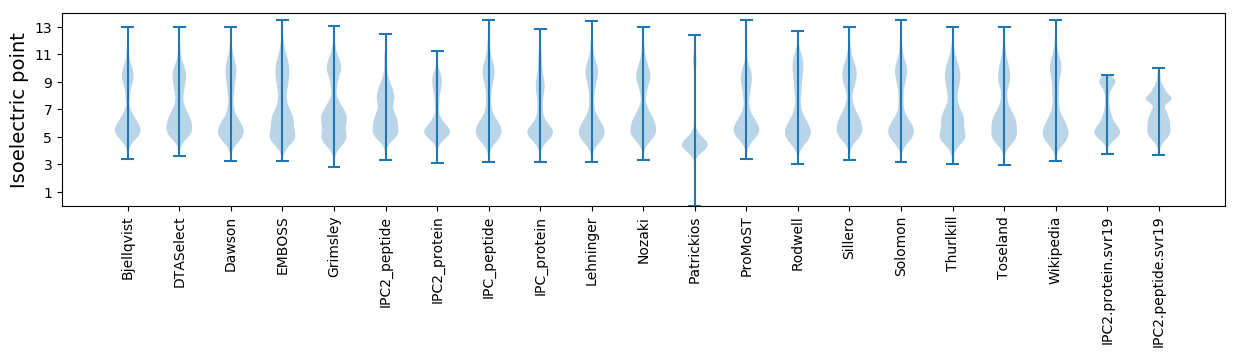
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A4FRD8|A0A2A4FRD8_9SPHN MFS transporter OS=Rhizorhabdus dicambivorans OX=1850238 GN=COO09_17390 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.61GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.61GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1542366 |
22 |
4544 |
317.6 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.227 ± 0.052 | 0.825 ± 0.011 |
5.958 ± 0.027 | 5.444 ± 0.036 |
3.529 ± 0.018 | 9.002 ± 0.057 |
2.049 ± 0.019 | 5.297 ± 0.02 |
2.892 ± 0.031 | 9.945 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.446 ± 0.016 | 2.454 ± 0.025 |
5.35 ± 0.025 | 3.038 ± 0.018 |
7.712 ± 0.041 | 5.3 ± 0.026 |
4.984 ± 0.029 | 6.924 ± 0.03 |
1.398 ± 0.016 | 2.227 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
