
Yongjia Tick Virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Uukuvirus; Yongjia uukuvirus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
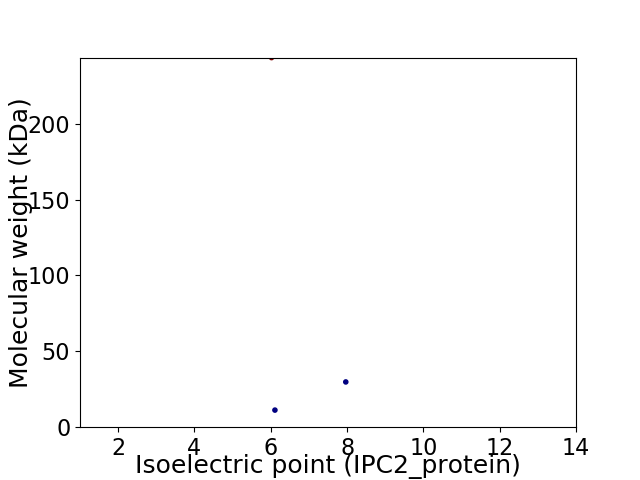
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
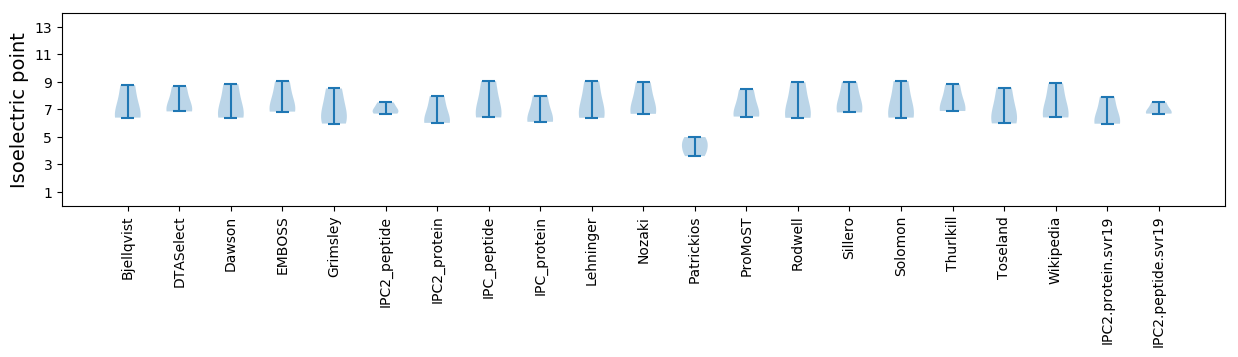
Summary statistics related to proteome-wise predictions
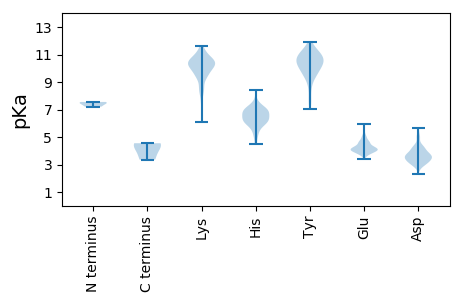
Protein with the lowest isoelectric point:
>tr|A0A0B5KS85|A0A0B5KS85_9VIRU Nonstructural protein OS=Yongjia Tick Virus 1 OX=1608145 GN=NSs PE=4 SV=1
MM1 pKa = 7.21RR2 pKa = 11.84TEE4 pKa = 3.7TMMDD8 pKa = 3.01KK9 pKa = 11.24GFISQLDD16 pKa = 3.66PLLCSRR22 pKa = 11.84LGDD25 pKa = 3.74LLKK28 pKa = 10.38QLRR31 pKa = 11.84ATEE34 pKa = 4.07VGRR37 pKa = 11.84RR38 pKa = 11.84SAWATPTRR46 pKa = 11.84QTNNDD51 pKa = 3.52EE52 pKa = 5.01NEE54 pKa = 4.3VCPLPPMCTPLVYY67 pKa = 10.28PSVLTVQLKK76 pKa = 8.54VWPLEE81 pKa = 4.22GPLVSSPPHH90 pKa = 6.26LCLCVCVGFVCGG102 pKa = 4.55
MM1 pKa = 7.21RR2 pKa = 11.84TEE4 pKa = 3.7TMMDD8 pKa = 3.01KK9 pKa = 11.24GFISQLDD16 pKa = 3.66PLLCSRR22 pKa = 11.84LGDD25 pKa = 3.74LLKK28 pKa = 10.38QLRR31 pKa = 11.84ATEE34 pKa = 4.07VGRR37 pKa = 11.84RR38 pKa = 11.84SAWATPTRR46 pKa = 11.84QTNNDD51 pKa = 3.52EE52 pKa = 5.01NEE54 pKa = 4.3VCPLPPMCTPLVYY67 pKa = 10.28PSVLTVQLKK76 pKa = 8.54VWPLEE81 pKa = 4.22GPLVSSPPHH90 pKa = 6.26LCLCVCVGFVCGG102 pKa = 4.55
Molecular weight: 11.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KS85|A0A0B5KS85_9VIRU Nonstructural protein OS=Yongjia Tick Virus 1 OX=1608145 GN=NSs PE=4 SV=1
MM1 pKa = 7.55AEE3 pKa = 4.64KK4 pKa = 10.19IRR6 pKa = 11.84FTGLPSLPDD15 pKa = 3.4VPTAADD21 pKa = 3.27WSRR24 pKa = 11.84VSFEE28 pKa = 4.07MGMTGAPEE36 pKa = 3.86VEE38 pKa = 5.08AITEE42 pKa = 3.93LSEE45 pKa = 3.86MFKK48 pKa = 10.64YY49 pKa = 10.36QGMDD53 pKa = 3.16PAVIIKK59 pKa = 10.29KK60 pKa = 9.67FATLGKK66 pKa = 8.92EE67 pKa = 4.05AHH69 pKa = 6.64RR70 pKa = 11.84NWLNDD75 pKa = 3.07AGFLIVLHH83 pKa = 6.69ICRR86 pKa = 11.84GTQVTKK92 pKa = 10.37IKK94 pKa = 9.81KK95 pKa = 7.59TVTPEE100 pKa = 3.41TATEE104 pKa = 4.02IEE106 pKa = 4.51TLVATYY112 pKa = 9.98KK113 pKa = 10.56IKK115 pKa = 10.59DD116 pKa = 3.75KK117 pKa = 11.3KK118 pKa = 10.53PVGDD122 pKa = 5.57DD123 pKa = 2.84ITLARR128 pKa = 11.84IALCFPLLTLRR139 pKa = 11.84QLSNFQDD146 pKa = 3.47HH147 pKa = 6.22LTVKK151 pKa = 8.76HH152 pKa = 5.7HH153 pKa = 6.64HH154 pKa = 5.89MEE156 pKa = 4.27EE157 pKa = 4.1LSAGYY162 pKa = 8.69PIQLMHH168 pKa = 6.76SAFASLITTTAGRR181 pKa = 11.84QQDD184 pKa = 3.96TQDD187 pKa = 3.95LLDD190 pKa = 3.37AHH192 pKa = 7.09RR193 pKa = 11.84LYY195 pKa = 10.92LVEE198 pKa = 4.0LTKK201 pKa = 10.9VINVGMKK208 pKa = 9.42GKK210 pKa = 7.82TVAEE214 pKa = 3.93IAEE217 pKa = 4.49SFEE220 pKa = 4.22QPLQAGFNSTFVSQSHH236 pKa = 6.13RR237 pKa = 11.84AASLRR242 pKa = 11.84KK243 pKa = 9.9LGVVDD248 pKa = 5.1GNGSPIAAVRR258 pKa = 11.84AAAQVYY264 pKa = 7.42RR265 pKa = 11.84TLKK268 pKa = 10.69ARR270 pKa = 11.84GG271 pKa = 3.36
MM1 pKa = 7.55AEE3 pKa = 4.64KK4 pKa = 10.19IRR6 pKa = 11.84FTGLPSLPDD15 pKa = 3.4VPTAADD21 pKa = 3.27WSRR24 pKa = 11.84VSFEE28 pKa = 4.07MGMTGAPEE36 pKa = 3.86VEE38 pKa = 5.08AITEE42 pKa = 3.93LSEE45 pKa = 3.86MFKK48 pKa = 10.64YY49 pKa = 10.36QGMDD53 pKa = 3.16PAVIIKK59 pKa = 10.29KK60 pKa = 9.67FATLGKK66 pKa = 8.92EE67 pKa = 4.05AHH69 pKa = 6.64RR70 pKa = 11.84NWLNDD75 pKa = 3.07AGFLIVLHH83 pKa = 6.69ICRR86 pKa = 11.84GTQVTKK92 pKa = 10.37IKK94 pKa = 9.81KK95 pKa = 7.59TVTPEE100 pKa = 3.41TATEE104 pKa = 4.02IEE106 pKa = 4.51TLVATYY112 pKa = 9.98KK113 pKa = 10.56IKK115 pKa = 10.59DD116 pKa = 3.75KK117 pKa = 11.3KK118 pKa = 10.53PVGDD122 pKa = 5.57DD123 pKa = 2.84ITLARR128 pKa = 11.84IALCFPLLTLRR139 pKa = 11.84QLSNFQDD146 pKa = 3.47HH147 pKa = 6.22LTVKK151 pKa = 8.76HH152 pKa = 5.7HH153 pKa = 6.64HH154 pKa = 5.89MEE156 pKa = 4.27EE157 pKa = 4.1LSAGYY162 pKa = 8.69PIQLMHH168 pKa = 6.76SAFASLITTTAGRR181 pKa = 11.84QQDD184 pKa = 3.96TQDD187 pKa = 3.95LLDD190 pKa = 3.37AHH192 pKa = 7.09RR193 pKa = 11.84LYY195 pKa = 10.92LVEE198 pKa = 4.0LTKK201 pKa = 10.9VINVGMKK208 pKa = 9.42GKK210 pKa = 7.82TVAEE214 pKa = 3.93IAEE217 pKa = 4.49SFEE220 pKa = 4.22QPLQAGFNSTFVSQSHH236 pKa = 6.13RR237 pKa = 11.84AASLRR242 pKa = 11.84KK243 pKa = 9.9LGVVDD248 pKa = 5.1GNGSPIAAVRR258 pKa = 11.84AAAQVYY264 pKa = 7.42RR265 pKa = 11.84TLKK268 pKa = 10.69ARR270 pKa = 11.84GG271 pKa = 3.36
Molecular weight: 29.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2511 |
102 |
2138 |
837.0 |
94.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.531 ± 1.415 | 1.513 ± 0.967 |
5.297 ± 0.501 | 7.527 ± 0.946 |
4.261 ± 0.489 | 5.297 ± 0.426 |
2.867 ± 0.337 | 5.536 ± 0.847 |
6.213 ± 0.594 | 9.478 ± 1.116 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.266 ± 0.133 | 3.067 ± 0.329 |
4.54 ± 1.164 | 3.266 ± 0.502 |
5.974 ± 0.447 | 8.244 ± 1.407 |
6.452 ± 1.042 | 6.252 ± 0.856 |
1.354 ± 0.21 | 3.067 ± 0.73 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
