
Bacillus caseinilyticus
Taxonomy: cellular organisms; Bacteria;
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
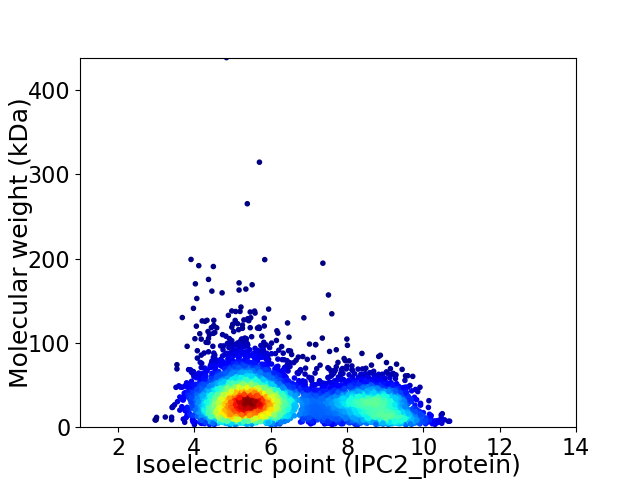
Virtual 2D-PAGE plot for 5197 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
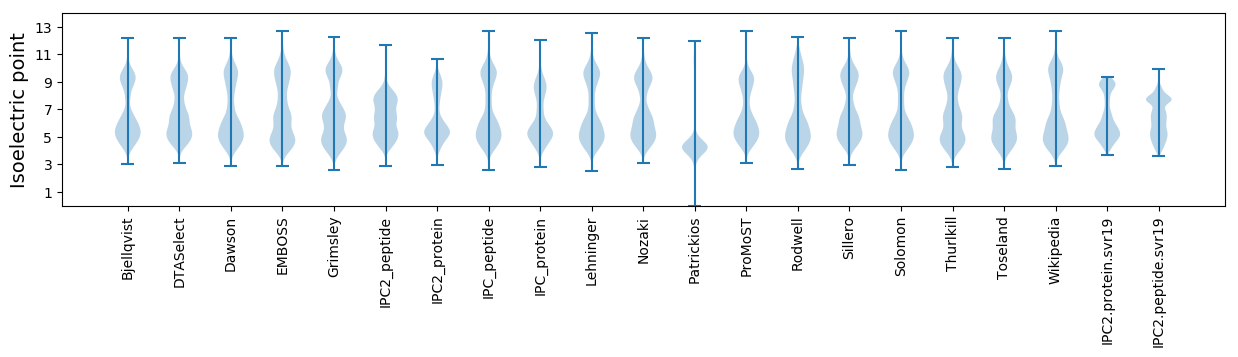
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3GMA5|A0A1H3GMA5_9BACI Uncharacterized protein OS=Bacillus caseinilyticus OX=1503961 GN=SAMN05421736_101218 PE=4 SV=1
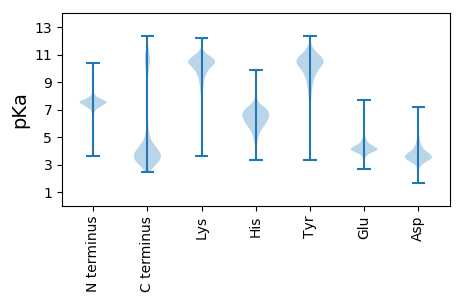
MM1 pKa = 7.84PIKK4 pKa = 10.26KK5 pKa = 9.81RR6 pKa = 11.84YY7 pKa = 9.4LFLASLFMTALLFGCSDD24 pKa = 3.42NSNNADD30 pKa = 4.36DD31 pKa = 4.57DD32 pKa = 4.25TGSQEE37 pKa = 4.01PVADD41 pKa = 4.36EE42 pKa = 4.85SSDD45 pKa = 3.98SADD48 pKa = 3.33SDD50 pKa = 3.7SGRR53 pKa = 11.84VLTDD57 pKa = 3.33AVGHH61 pKa = 5.98EE62 pKa = 4.48VTIPDD67 pKa = 3.9APEE70 pKa = 4.52KK71 pKa = 10.55IIASYY76 pKa = 11.13LEE78 pKa = 4.9DD79 pKa = 3.81YY80 pKa = 10.96LVALDD85 pKa = 3.99VTPAAQWSILDD96 pKa = 3.81GQGLQDD102 pKa = 3.76YY103 pKa = 10.07LQDD106 pKa = 3.72YY107 pKa = 8.65LQAVPPIPYY116 pKa = 8.51EE117 pKa = 3.82LPYY120 pKa = 10.91EE121 pKa = 4.42SVTSFSPDD129 pKa = 3.12LLLISSAEE137 pKa = 3.89AVDD140 pKa = 3.76DD141 pKa = 4.48GKK143 pKa = 11.29YY144 pKa = 9.51EE145 pKa = 4.1QYY147 pKa = 11.45SKK149 pKa = 10.72IAPTYY154 pKa = 10.76VVGADD159 pKa = 3.61NNSDD163 pKa = 2.81WRR165 pKa = 11.84EE166 pKa = 3.86NLLTIAEE173 pKa = 4.25VLDD176 pKa = 4.45LEE178 pKa = 5.25NKK180 pKa = 9.34AQQLLASYY188 pKa = 10.16DD189 pKa = 3.75EE190 pKa = 4.49KK191 pKa = 11.42AQAAKK196 pKa = 10.83EE197 pKa = 4.06EE198 pKa = 4.08IQAAIGTDD206 pKa = 3.2ATAAALWLVADD217 pKa = 3.9SFFIVSEE224 pKa = 4.23NVSSGAVLYY233 pKa = 9.84EE234 pKa = 4.11DD235 pKa = 5.33LGIAVPDD242 pKa = 3.69VVKK245 pKa = 10.67EE246 pKa = 4.0ISGDD250 pKa = 3.57AAANWSPISLEE261 pKa = 4.15EE262 pKa = 4.03IATLEE267 pKa = 4.12ADD269 pKa = 3.8HH270 pKa = 6.91LFLINSDD277 pKa = 3.71NASGAEE283 pKa = 3.95MLNDD287 pKa = 4.31PVWQSIPAVKK297 pKa = 10.27NGQVYY302 pKa = 9.94EE303 pKa = 4.19YY304 pKa = 10.89GPEE307 pKa = 4.16TSWLYY312 pKa = 9.9TGPIASAQMIDD323 pKa = 3.74HH324 pKa = 5.86VVEE327 pKa = 4.42SLVKK331 pKa = 10.51
MM1 pKa = 7.84PIKK4 pKa = 10.26KK5 pKa = 9.81RR6 pKa = 11.84YY7 pKa = 9.4LFLASLFMTALLFGCSDD24 pKa = 3.42NSNNADD30 pKa = 4.36DD31 pKa = 4.57DD32 pKa = 4.25TGSQEE37 pKa = 4.01PVADD41 pKa = 4.36EE42 pKa = 4.85SSDD45 pKa = 3.98SADD48 pKa = 3.33SDD50 pKa = 3.7SGRR53 pKa = 11.84VLTDD57 pKa = 3.33AVGHH61 pKa = 5.98EE62 pKa = 4.48VTIPDD67 pKa = 3.9APEE70 pKa = 4.52KK71 pKa = 10.55IIASYY76 pKa = 11.13LEE78 pKa = 4.9DD79 pKa = 3.81YY80 pKa = 10.96LVALDD85 pKa = 3.99VTPAAQWSILDD96 pKa = 3.81GQGLQDD102 pKa = 3.76YY103 pKa = 10.07LQDD106 pKa = 3.72YY107 pKa = 8.65LQAVPPIPYY116 pKa = 8.51EE117 pKa = 3.82LPYY120 pKa = 10.91EE121 pKa = 4.42SVTSFSPDD129 pKa = 3.12LLLISSAEE137 pKa = 3.89AVDD140 pKa = 3.76DD141 pKa = 4.48GKK143 pKa = 11.29YY144 pKa = 9.51EE145 pKa = 4.1QYY147 pKa = 11.45SKK149 pKa = 10.72IAPTYY154 pKa = 10.76VVGADD159 pKa = 3.61NNSDD163 pKa = 2.81WRR165 pKa = 11.84EE166 pKa = 3.86NLLTIAEE173 pKa = 4.25VLDD176 pKa = 4.45LEE178 pKa = 5.25NKK180 pKa = 9.34AQQLLASYY188 pKa = 10.16DD189 pKa = 3.75EE190 pKa = 4.49KK191 pKa = 11.42AQAAKK196 pKa = 10.83EE197 pKa = 4.06EE198 pKa = 4.08IQAAIGTDD206 pKa = 3.2ATAAALWLVADD217 pKa = 3.9SFFIVSEE224 pKa = 4.23NVSSGAVLYY233 pKa = 9.84EE234 pKa = 4.11DD235 pKa = 5.33LGIAVPDD242 pKa = 3.69VVKK245 pKa = 10.67EE246 pKa = 4.0ISGDD250 pKa = 3.57AAANWSPISLEE261 pKa = 4.15EE262 pKa = 4.03IATLEE267 pKa = 4.12ADD269 pKa = 3.8HH270 pKa = 6.91LFLINSDD277 pKa = 3.71NASGAEE283 pKa = 3.95MLNDD287 pKa = 4.31PVWQSIPAVKK297 pKa = 10.27NGQVYY302 pKa = 9.94EE303 pKa = 4.19YY304 pKa = 10.89GPEE307 pKa = 4.16TSWLYY312 pKa = 9.9TGPIASAQMIDD323 pKa = 3.74HH324 pKa = 5.86VVEE327 pKa = 4.42SLVKK331 pKa = 10.51
Molecular weight: 35.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3TLQ3|A0A1H3TLQ3_9BACI Uncharacterized protein OS=Bacillus caseinilyticus OX=1503961 GN=SAMN05421736_11530 PE=4 SV=1
MM1 pKa = 7.2ITQTTVPDD9 pKa = 3.51VCFVRR14 pKa = 11.84PRR16 pKa = 11.84LTYY19 pKa = 10.48KK20 pKa = 10.76RR21 pKa = 11.84IAGCLLLAAVPIRR34 pKa = 11.84LVPLLLGSGTSFFPLKK50 pKa = 10.52QSS52 pKa = 3.18
MM1 pKa = 7.2ITQTTVPDD9 pKa = 3.51VCFVRR14 pKa = 11.84PRR16 pKa = 11.84LTYY19 pKa = 10.48KK20 pKa = 10.76RR21 pKa = 11.84IAGCLLLAAVPIRR34 pKa = 11.84LVPLLLGSGTSFFPLKK50 pKa = 10.52QSS52 pKa = 3.18
Molecular weight: 5.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1567055 |
25 |
3823 |
301.5 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.698 ± 0.035 | 0.786 ± 0.012 |
5.112 ± 0.028 | 7.512 ± 0.042 |
4.576 ± 0.028 | 6.898 ± 0.029 |
2.198 ± 0.019 | 7.309 ± 0.032 |
6.122 ± 0.033 | 9.797 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.018 | 4.116 ± 0.025 |
3.697 ± 0.022 | 3.844 ± 0.022 |
4.479 ± 0.029 | 5.845 ± 0.022 |
5.414 ± 0.023 | 7.065 ± 0.027 |
1.136 ± 0.014 | 3.616 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
