
Rhizoctonia oryzae-sativae mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
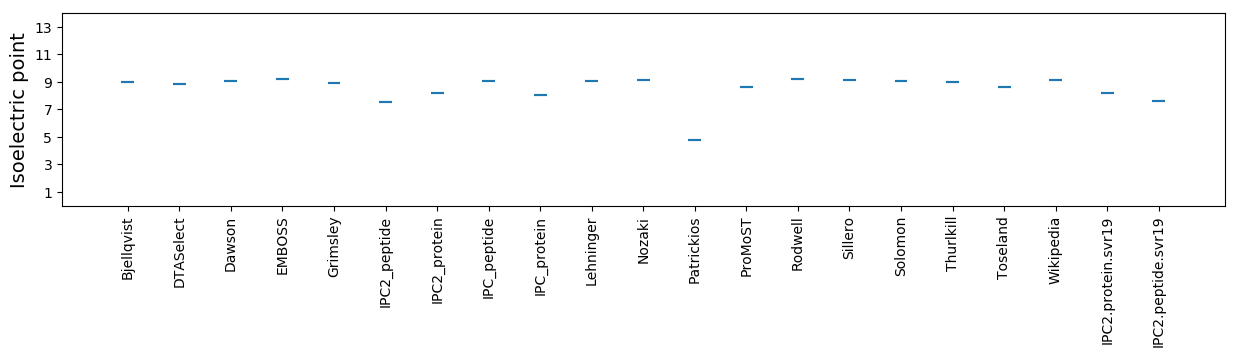
Average proteome isoelectric point is 8.17
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A166H5D4|A0A166H5D4_9VIRU Putative RNA-dependent RNA polymerase OS=Rhizoctonia oryzae-sativae mitovirus 1 OX=1837089 PE=4 SV=1
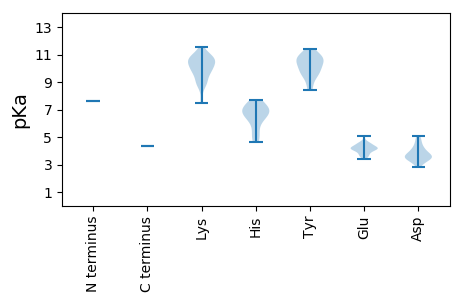
MM1 pKa = 7.6KK2 pKa = 10.24IINTTINNKK11 pKa = 8.87TNTNFLSFSSGYY23 pKa = 9.77SFSSFSPNATNSTTVKK39 pKa = 10.44DD40 pKa = 3.89LRR42 pKa = 11.84QSIVPGINVVRR53 pKa = 11.84NSTRR57 pKa = 11.84LARR60 pKa = 11.84SIALLTAYY68 pKa = 9.65IEE70 pKa = 4.1IQLRR74 pKa = 11.84DD75 pKa = 3.73LDD77 pKa = 3.94DD78 pKa = 3.63TSRR81 pKa = 11.84MVLKK85 pKa = 10.75QFMTEE90 pKa = 4.16LEE92 pKa = 4.38SMYY95 pKa = 9.51HH96 pKa = 5.39TNSSNFVAASKK107 pKa = 10.81QLGNWLKK114 pKa = 10.8VNLFKK119 pKa = 10.78KK120 pKa = 10.46VRR122 pKa = 11.84LQGDD126 pKa = 3.72VKK128 pKa = 10.62DD129 pKa = 4.17VHH131 pKa = 6.48WNAHH135 pKa = 4.61VSCPNQLVGVIAEE148 pKa = 4.04FHH150 pKa = 6.57RR151 pKa = 11.84IEE153 pKa = 4.6KK154 pKa = 8.92GTLKK158 pKa = 9.22TQYY161 pKa = 10.69LRR163 pKa = 11.84ALLSILNLYY172 pKa = 10.17EE173 pKa = 5.06SITIPTNPSLDD184 pKa = 4.13TITKK188 pKa = 9.18PSNVDD193 pKa = 2.94QTQISSQEE201 pKa = 3.61ILTALEE207 pKa = 3.91TLGIDD212 pKa = 3.64VAAVTNDD219 pKa = 3.42FKK221 pKa = 11.55FMNQSPTSHH230 pKa = 6.61SSSSAGPNGQAVWYY244 pKa = 9.61SHH246 pKa = 7.04LDD248 pKa = 4.03AKK250 pKa = 11.02ALWQDD255 pKa = 3.2EE256 pKa = 4.37TLKK259 pKa = 10.26MRR261 pKa = 11.84FNSLATILEE270 pKa = 4.21RR271 pKa = 11.84EE272 pKa = 4.24DD273 pKa = 4.87LKK275 pKa = 10.62TLLKK279 pKa = 10.62EE280 pKa = 4.25SAGLPKK286 pKa = 10.24LQKK289 pKa = 10.74ANRR292 pKa = 11.84NKK294 pKa = 9.87PNQPKK299 pKa = 7.62QHH301 pKa = 6.72KK302 pKa = 9.8IYY304 pKa = 10.8EE305 pKa = 4.55KK306 pKa = 10.38GNKK309 pKa = 7.49TRR311 pKa = 11.84VIAILDD317 pKa = 3.79YY318 pKa = 8.79FTQEE322 pKa = 3.55ILTPFHH328 pKa = 7.19DD329 pKa = 4.69LVASILKK336 pKa = 9.65KK337 pKa = 10.22IEE339 pKa = 3.64MDD341 pKa = 3.48GTFDD345 pKa = 3.33QDD347 pKa = 4.9KK348 pKa = 9.12IARR351 pKa = 11.84WVKK354 pKa = 10.41SKK356 pKa = 8.99TAQSCVSLYY365 pKa = 10.91SYY367 pKa = 11.43DD368 pKa = 3.31LTAATDD374 pKa = 4.0RR375 pKa = 11.84LPVWLQRR382 pKa = 11.84RR383 pKa = 11.84IIEE386 pKa = 4.17CLIKK390 pKa = 10.01IDD392 pKa = 4.24NFGLNWQLLLTDD404 pKa = 4.36RR405 pKa = 11.84AFDD408 pKa = 3.74NPIGEE413 pKa = 4.23PVKK416 pKa = 10.86YY417 pKa = 10.31CVGQPMGAKK426 pKa = 9.91SSFPMLGLTHH436 pKa = 6.91HH437 pKa = 7.24IIVQIAAKK445 pKa = 8.89RR446 pKa = 11.84TGFQGLFTEE455 pKa = 4.55YY456 pKa = 10.75VILGDD461 pKa = 5.11DD462 pKa = 3.2ICIADD467 pKa = 4.55DD468 pKa = 4.45LVAAKK473 pKa = 9.65YY474 pKa = 10.3KK475 pKa = 11.09EE476 pKa = 3.88MMIEE480 pKa = 4.14LGLTISEE487 pKa = 4.52HH488 pKa = 5.04KK489 pKa = 9.66TIVSTVEE496 pKa = 3.65TSIDD500 pKa = 3.54PVAEE504 pKa = 3.41ICKK507 pKa = 9.69RR508 pKa = 11.84VFIGGVEE515 pKa = 4.09VTPLPMKK522 pKa = 10.45LAANVTEE529 pKa = 4.13NNDD532 pKa = 3.06LFFQFRR538 pKa = 11.84EE539 pKa = 4.16KK540 pKa = 10.72LSEE543 pKa = 4.11RR544 pKa = 11.84GLIYY548 pKa = 10.97NPADD552 pKa = 2.84WKK554 pKa = 11.05YY555 pKa = 10.29FVAACVSNHH564 pKa = 6.15RR565 pKa = 11.84SLEE568 pKa = 4.11KK569 pKa = 10.16IGLYY573 pKa = 10.76NSMPLWLSGFKK584 pKa = 10.16QSLPIGTVGNNNFEE598 pKa = 4.53DD599 pKa = 3.63WLKK602 pKa = 11.45AGMTEE607 pKa = 4.31DD608 pKa = 3.71YY609 pKa = 10.79LRR611 pKa = 11.84EE612 pKa = 3.84VWTFTVLQEE621 pKa = 3.84QFKK624 pKa = 10.71RR625 pKa = 11.84ISTLIQSAQDD635 pKa = 3.38TFQTISKK642 pKa = 9.8GIRR645 pKa = 11.84LGDD648 pKa = 3.41AVYY651 pKa = 10.1IVPDD655 pKa = 3.55TGIEE659 pKa = 4.09LPVTKK664 pKa = 10.31FDD666 pKa = 3.5IQKK669 pKa = 8.83MNEE672 pKa = 3.69WNILNIPHH680 pKa = 7.67PARR683 pKa = 11.84FVMLGEE689 pKa = 4.25VNRR692 pKa = 11.84VATLFSQLSSIRR704 pKa = 11.84GPSLMTKK711 pKa = 8.85LTHH714 pKa = 7.19NIVDD718 pKa = 3.59NLKK721 pKa = 10.45FSVLEE726 pKa = 4.62MINDD730 pKa = 3.32MDD732 pKa = 4.05NYY734 pKa = 9.42KK735 pKa = 10.89ARR737 pKa = 11.84VDD739 pKa = 3.65RR740 pKa = 11.84QLLEE744 pKa = 4.33KK745 pKa = 9.62MISNIRR751 pKa = 11.84MIIRR755 pKa = 11.84AEE757 pKa = 4.1NKK759 pKa = 8.45TVTYY763 pKa = 8.42TVKK766 pKa = 9.76LTPLGIVWVLRR777 pKa = 11.84LSHH780 pKa = 6.98LGSCTLFRR788 pKa = 11.84STSAIPTTLRR798 pKa = 11.84DD799 pKa = 3.44AEE801 pKa = 4.41LKK803 pKa = 9.79FKK805 pKa = 10.88RR806 pKa = 11.84MNSGNSQIASIFDD819 pKa = 3.48TT820 pKa = 4.34
MM1 pKa = 7.6KK2 pKa = 10.24IINTTINNKK11 pKa = 8.87TNTNFLSFSSGYY23 pKa = 9.77SFSSFSPNATNSTTVKK39 pKa = 10.44DD40 pKa = 3.89LRR42 pKa = 11.84QSIVPGINVVRR53 pKa = 11.84NSTRR57 pKa = 11.84LARR60 pKa = 11.84SIALLTAYY68 pKa = 9.65IEE70 pKa = 4.1IQLRR74 pKa = 11.84DD75 pKa = 3.73LDD77 pKa = 3.94DD78 pKa = 3.63TSRR81 pKa = 11.84MVLKK85 pKa = 10.75QFMTEE90 pKa = 4.16LEE92 pKa = 4.38SMYY95 pKa = 9.51HH96 pKa = 5.39TNSSNFVAASKK107 pKa = 10.81QLGNWLKK114 pKa = 10.8VNLFKK119 pKa = 10.78KK120 pKa = 10.46VRR122 pKa = 11.84LQGDD126 pKa = 3.72VKK128 pKa = 10.62DD129 pKa = 4.17VHH131 pKa = 6.48WNAHH135 pKa = 4.61VSCPNQLVGVIAEE148 pKa = 4.04FHH150 pKa = 6.57RR151 pKa = 11.84IEE153 pKa = 4.6KK154 pKa = 8.92GTLKK158 pKa = 9.22TQYY161 pKa = 10.69LRR163 pKa = 11.84ALLSILNLYY172 pKa = 10.17EE173 pKa = 5.06SITIPTNPSLDD184 pKa = 4.13TITKK188 pKa = 9.18PSNVDD193 pKa = 2.94QTQISSQEE201 pKa = 3.61ILTALEE207 pKa = 3.91TLGIDD212 pKa = 3.64VAAVTNDD219 pKa = 3.42FKK221 pKa = 11.55FMNQSPTSHH230 pKa = 6.61SSSSAGPNGQAVWYY244 pKa = 9.61SHH246 pKa = 7.04LDD248 pKa = 4.03AKK250 pKa = 11.02ALWQDD255 pKa = 3.2EE256 pKa = 4.37TLKK259 pKa = 10.26MRR261 pKa = 11.84FNSLATILEE270 pKa = 4.21RR271 pKa = 11.84EE272 pKa = 4.24DD273 pKa = 4.87LKK275 pKa = 10.62TLLKK279 pKa = 10.62EE280 pKa = 4.25SAGLPKK286 pKa = 10.24LQKK289 pKa = 10.74ANRR292 pKa = 11.84NKK294 pKa = 9.87PNQPKK299 pKa = 7.62QHH301 pKa = 6.72KK302 pKa = 9.8IYY304 pKa = 10.8EE305 pKa = 4.55KK306 pKa = 10.38GNKK309 pKa = 7.49TRR311 pKa = 11.84VIAILDD317 pKa = 3.79YY318 pKa = 8.79FTQEE322 pKa = 3.55ILTPFHH328 pKa = 7.19DD329 pKa = 4.69LVASILKK336 pKa = 9.65KK337 pKa = 10.22IEE339 pKa = 3.64MDD341 pKa = 3.48GTFDD345 pKa = 3.33QDD347 pKa = 4.9KK348 pKa = 9.12IARR351 pKa = 11.84WVKK354 pKa = 10.41SKK356 pKa = 8.99TAQSCVSLYY365 pKa = 10.91SYY367 pKa = 11.43DD368 pKa = 3.31LTAATDD374 pKa = 4.0RR375 pKa = 11.84LPVWLQRR382 pKa = 11.84RR383 pKa = 11.84IIEE386 pKa = 4.17CLIKK390 pKa = 10.01IDD392 pKa = 4.24NFGLNWQLLLTDD404 pKa = 4.36RR405 pKa = 11.84AFDD408 pKa = 3.74NPIGEE413 pKa = 4.23PVKK416 pKa = 10.86YY417 pKa = 10.31CVGQPMGAKK426 pKa = 9.91SSFPMLGLTHH436 pKa = 6.91HH437 pKa = 7.24IIVQIAAKK445 pKa = 8.89RR446 pKa = 11.84TGFQGLFTEE455 pKa = 4.55YY456 pKa = 10.75VILGDD461 pKa = 5.11DD462 pKa = 3.2ICIADD467 pKa = 4.55DD468 pKa = 4.45LVAAKK473 pKa = 9.65YY474 pKa = 10.3KK475 pKa = 11.09EE476 pKa = 3.88MMIEE480 pKa = 4.14LGLTISEE487 pKa = 4.52HH488 pKa = 5.04KK489 pKa = 9.66TIVSTVEE496 pKa = 3.65TSIDD500 pKa = 3.54PVAEE504 pKa = 3.41ICKK507 pKa = 9.69RR508 pKa = 11.84VFIGGVEE515 pKa = 4.09VTPLPMKK522 pKa = 10.45LAANVTEE529 pKa = 4.13NNDD532 pKa = 3.06LFFQFRR538 pKa = 11.84EE539 pKa = 4.16KK540 pKa = 10.72LSEE543 pKa = 4.11RR544 pKa = 11.84GLIYY548 pKa = 10.97NPADD552 pKa = 2.84WKK554 pKa = 11.05YY555 pKa = 10.29FVAACVSNHH564 pKa = 6.15RR565 pKa = 11.84SLEE568 pKa = 4.11KK569 pKa = 10.16IGLYY573 pKa = 10.76NSMPLWLSGFKK584 pKa = 10.16QSLPIGTVGNNNFEE598 pKa = 4.53DD599 pKa = 3.63WLKK602 pKa = 11.45AGMTEE607 pKa = 4.31DD608 pKa = 3.71YY609 pKa = 10.79LRR611 pKa = 11.84EE612 pKa = 3.84VWTFTVLQEE621 pKa = 3.84QFKK624 pKa = 10.71RR625 pKa = 11.84ISTLIQSAQDD635 pKa = 3.38TFQTISKK642 pKa = 9.8GIRR645 pKa = 11.84LGDD648 pKa = 3.41AVYY651 pKa = 10.1IVPDD655 pKa = 3.55TGIEE659 pKa = 4.09LPVTKK664 pKa = 10.31FDD666 pKa = 3.5IQKK669 pKa = 8.83MNEE672 pKa = 3.69WNILNIPHH680 pKa = 7.67PARR683 pKa = 11.84FVMLGEE689 pKa = 4.25VNRR692 pKa = 11.84VATLFSQLSSIRR704 pKa = 11.84GPSLMTKK711 pKa = 8.85LTHH714 pKa = 7.19NIVDD718 pKa = 3.59NLKK721 pKa = 10.45FSVLEE726 pKa = 4.62MINDD730 pKa = 3.32MDD732 pKa = 4.05NYY734 pKa = 9.42KK735 pKa = 10.89ARR737 pKa = 11.84VDD739 pKa = 3.65RR740 pKa = 11.84QLLEE744 pKa = 4.33KK745 pKa = 9.62MISNIRR751 pKa = 11.84MIIRR755 pKa = 11.84AEE757 pKa = 4.1NKK759 pKa = 8.45TVTYY763 pKa = 8.42TVKK766 pKa = 9.76LTPLGIVWVLRR777 pKa = 11.84LSHH780 pKa = 6.98LGSCTLFRR788 pKa = 11.84STSAIPTTLRR798 pKa = 11.84DD799 pKa = 3.44AEE801 pKa = 4.41LKK803 pKa = 9.79FKK805 pKa = 10.88RR806 pKa = 11.84MNSGNSQIASIFDD819 pKa = 3.48TT820 pKa = 4.34
Molecular weight: 92.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166H5D4|A0A166H5D4_9VIRU Putative RNA-dependent RNA polymerase OS=Rhizoctonia oryzae-sativae mitovirus 1 OX=1837089 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.24IINTTINNKK11 pKa = 8.87TNTNFLSFSSGYY23 pKa = 9.77SFSSFSPNATNSTTVKK39 pKa = 10.44DD40 pKa = 3.89LRR42 pKa = 11.84QSIVPGINVVRR53 pKa = 11.84NSTRR57 pKa = 11.84LARR60 pKa = 11.84SIALLTAYY68 pKa = 9.65IEE70 pKa = 4.1IQLRR74 pKa = 11.84DD75 pKa = 3.73LDD77 pKa = 3.94DD78 pKa = 3.63TSRR81 pKa = 11.84MVLKK85 pKa = 10.75QFMTEE90 pKa = 4.16LEE92 pKa = 4.38SMYY95 pKa = 9.51HH96 pKa = 5.39TNSSNFVAASKK107 pKa = 10.81QLGNWLKK114 pKa = 10.8VNLFKK119 pKa = 10.78KK120 pKa = 10.46VRR122 pKa = 11.84LQGDD126 pKa = 3.72VKK128 pKa = 10.62DD129 pKa = 4.17VHH131 pKa = 6.48WNAHH135 pKa = 4.61VSCPNQLVGVIAEE148 pKa = 4.04FHH150 pKa = 6.57RR151 pKa = 11.84IEE153 pKa = 4.6KK154 pKa = 8.92GTLKK158 pKa = 9.22TQYY161 pKa = 10.69LRR163 pKa = 11.84ALLSILNLYY172 pKa = 10.17EE173 pKa = 5.06SITIPTNPSLDD184 pKa = 4.13TITKK188 pKa = 9.18PSNVDD193 pKa = 2.94QTQISSQEE201 pKa = 3.61ILTALEE207 pKa = 3.91TLGIDD212 pKa = 3.64VAAVTNDD219 pKa = 3.42FKK221 pKa = 11.55FMNQSPTSHH230 pKa = 6.61SSSSAGPNGQAVWYY244 pKa = 9.61SHH246 pKa = 7.04LDD248 pKa = 4.03AKK250 pKa = 11.02ALWQDD255 pKa = 3.2EE256 pKa = 4.37TLKK259 pKa = 10.26MRR261 pKa = 11.84FNSLATILEE270 pKa = 4.21RR271 pKa = 11.84EE272 pKa = 4.24DD273 pKa = 4.87LKK275 pKa = 10.62TLLKK279 pKa = 10.62EE280 pKa = 4.25SAGLPKK286 pKa = 10.24LQKK289 pKa = 10.74ANRR292 pKa = 11.84NKK294 pKa = 9.87PNQPKK299 pKa = 7.62QHH301 pKa = 6.72KK302 pKa = 9.8IYY304 pKa = 10.8EE305 pKa = 4.55KK306 pKa = 10.38GNKK309 pKa = 7.49TRR311 pKa = 11.84VIAILDD317 pKa = 3.79YY318 pKa = 8.79FTQEE322 pKa = 3.55ILTPFHH328 pKa = 7.19DD329 pKa = 4.69LVASILKK336 pKa = 9.65KK337 pKa = 10.22IEE339 pKa = 3.64MDD341 pKa = 3.48GTFDD345 pKa = 3.33QDD347 pKa = 4.9KK348 pKa = 9.12IARR351 pKa = 11.84WVKK354 pKa = 10.41SKK356 pKa = 8.99TAQSCVSLYY365 pKa = 10.91SYY367 pKa = 11.43DD368 pKa = 3.31LTAATDD374 pKa = 4.0RR375 pKa = 11.84LPVWLQRR382 pKa = 11.84RR383 pKa = 11.84IIEE386 pKa = 4.17CLIKK390 pKa = 10.01IDD392 pKa = 4.24NFGLNWQLLLTDD404 pKa = 4.36RR405 pKa = 11.84AFDD408 pKa = 3.74NPIGEE413 pKa = 4.23PVKK416 pKa = 10.86YY417 pKa = 10.31CVGQPMGAKK426 pKa = 9.91SSFPMLGLTHH436 pKa = 6.91HH437 pKa = 7.24IIVQIAAKK445 pKa = 8.89RR446 pKa = 11.84TGFQGLFTEE455 pKa = 4.55YY456 pKa = 10.75VILGDD461 pKa = 5.11DD462 pKa = 3.2ICIADD467 pKa = 4.55DD468 pKa = 4.45LVAAKK473 pKa = 9.65YY474 pKa = 10.3KK475 pKa = 11.09EE476 pKa = 3.88MMIEE480 pKa = 4.14LGLTISEE487 pKa = 4.52HH488 pKa = 5.04KK489 pKa = 9.66TIVSTVEE496 pKa = 3.65TSIDD500 pKa = 3.54PVAEE504 pKa = 3.41ICKK507 pKa = 9.69RR508 pKa = 11.84VFIGGVEE515 pKa = 4.09VTPLPMKK522 pKa = 10.45LAANVTEE529 pKa = 4.13NNDD532 pKa = 3.06LFFQFRR538 pKa = 11.84EE539 pKa = 4.16KK540 pKa = 10.72LSEE543 pKa = 4.11RR544 pKa = 11.84GLIYY548 pKa = 10.97NPADD552 pKa = 2.84WKK554 pKa = 11.05YY555 pKa = 10.29FVAACVSNHH564 pKa = 6.15RR565 pKa = 11.84SLEE568 pKa = 4.11KK569 pKa = 10.16IGLYY573 pKa = 10.76NSMPLWLSGFKK584 pKa = 10.16QSLPIGTVGNNNFEE598 pKa = 4.53DD599 pKa = 3.63WLKK602 pKa = 11.45AGMTEE607 pKa = 4.31DD608 pKa = 3.71YY609 pKa = 10.79LRR611 pKa = 11.84EE612 pKa = 3.84VWTFTVLQEE621 pKa = 3.84QFKK624 pKa = 10.71RR625 pKa = 11.84ISTLIQSAQDD635 pKa = 3.38TFQTISKK642 pKa = 9.8GIRR645 pKa = 11.84LGDD648 pKa = 3.41AVYY651 pKa = 10.1IVPDD655 pKa = 3.55TGIEE659 pKa = 4.09LPVTKK664 pKa = 10.31FDD666 pKa = 3.5IQKK669 pKa = 8.83MNEE672 pKa = 3.69WNILNIPHH680 pKa = 7.67PARR683 pKa = 11.84FVMLGEE689 pKa = 4.25VNRR692 pKa = 11.84VATLFSQLSSIRR704 pKa = 11.84GPSLMTKK711 pKa = 8.85LTHH714 pKa = 7.19NIVDD718 pKa = 3.59NLKK721 pKa = 10.45FSVLEE726 pKa = 4.62MINDD730 pKa = 3.32MDD732 pKa = 4.05NYY734 pKa = 9.42KK735 pKa = 10.89ARR737 pKa = 11.84VDD739 pKa = 3.65RR740 pKa = 11.84QLLEE744 pKa = 4.33KK745 pKa = 9.62MISNIRR751 pKa = 11.84MIIRR755 pKa = 11.84AEE757 pKa = 4.1NKK759 pKa = 8.45TVTYY763 pKa = 8.42TVKK766 pKa = 9.76LTPLGIVWVLRR777 pKa = 11.84LSHH780 pKa = 6.98LGSCTLFRR788 pKa = 11.84STSAIPTTLRR798 pKa = 11.84DD799 pKa = 3.44AEE801 pKa = 4.41LKK803 pKa = 9.79FKK805 pKa = 10.88RR806 pKa = 11.84MNSGNSQIASIFDD819 pKa = 3.48TT820 pKa = 4.34
MM1 pKa = 7.6KK2 pKa = 10.24IINTTINNKK11 pKa = 8.87TNTNFLSFSSGYY23 pKa = 9.77SFSSFSPNATNSTTVKK39 pKa = 10.44DD40 pKa = 3.89LRR42 pKa = 11.84QSIVPGINVVRR53 pKa = 11.84NSTRR57 pKa = 11.84LARR60 pKa = 11.84SIALLTAYY68 pKa = 9.65IEE70 pKa = 4.1IQLRR74 pKa = 11.84DD75 pKa = 3.73LDD77 pKa = 3.94DD78 pKa = 3.63TSRR81 pKa = 11.84MVLKK85 pKa = 10.75QFMTEE90 pKa = 4.16LEE92 pKa = 4.38SMYY95 pKa = 9.51HH96 pKa = 5.39TNSSNFVAASKK107 pKa = 10.81QLGNWLKK114 pKa = 10.8VNLFKK119 pKa = 10.78KK120 pKa = 10.46VRR122 pKa = 11.84LQGDD126 pKa = 3.72VKK128 pKa = 10.62DD129 pKa = 4.17VHH131 pKa = 6.48WNAHH135 pKa = 4.61VSCPNQLVGVIAEE148 pKa = 4.04FHH150 pKa = 6.57RR151 pKa = 11.84IEE153 pKa = 4.6KK154 pKa = 8.92GTLKK158 pKa = 9.22TQYY161 pKa = 10.69LRR163 pKa = 11.84ALLSILNLYY172 pKa = 10.17EE173 pKa = 5.06SITIPTNPSLDD184 pKa = 4.13TITKK188 pKa = 9.18PSNVDD193 pKa = 2.94QTQISSQEE201 pKa = 3.61ILTALEE207 pKa = 3.91TLGIDD212 pKa = 3.64VAAVTNDD219 pKa = 3.42FKK221 pKa = 11.55FMNQSPTSHH230 pKa = 6.61SSSSAGPNGQAVWYY244 pKa = 9.61SHH246 pKa = 7.04LDD248 pKa = 4.03AKK250 pKa = 11.02ALWQDD255 pKa = 3.2EE256 pKa = 4.37TLKK259 pKa = 10.26MRR261 pKa = 11.84FNSLATILEE270 pKa = 4.21RR271 pKa = 11.84EE272 pKa = 4.24DD273 pKa = 4.87LKK275 pKa = 10.62TLLKK279 pKa = 10.62EE280 pKa = 4.25SAGLPKK286 pKa = 10.24LQKK289 pKa = 10.74ANRR292 pKa = 11.84NKK294 pKa = 9.87PNQPKK299 pKa = 7.62QHH301 pKa = 6.72KK302 pKa = 9.8IYY304 pKa = 10.8EE305 pKa = 4.55KK306 pKa = 10.38GNKK309 pKa = 7.49TRR311 pKa = 11.84VIAILDD317 pKa = 3.79YY318 pKa = 8.79FTQEE322 pKa = 3.55ILTPFHH328 pKa = 7.19DD329 pKa = 4.69LVASILKK336 pKa = 9.65KK337 pKa = 10.22IEE339 pKa = 3.64MDD341 pKa = 3.48GTFDD345 pKa = 3.33QDD347 pKa = 4.9KK348 pKa = 9.12IARR351 pKa = 11.84WVKK354 pKa = 10.41SKK356 pKa = 8.99TAQSCVSLYY365 pKa = 10.91SYY367 pKa = 11.43DD368 pKa = 3.31LTAATDD374 pKa = 4.0RR375 pKa = 11.84LPVWLQRR382 pKa = 11.84RR383 pKa = 11.84IIEE386 pKa = 4.17CLIKK390 pKa = 10.01IDD392 pKa = 4.24NFGLNWQLLLTDD404 pKa = 4.36RR405 pKa = 11.84AFDD408 pKa = 3.74NPIGEE413 pKa = 4.23PVKK416 pKa = 10.86YY417 pKa = 10.31CVGQPMGAKK426 pKa = 9.91SSFPMLGLTHH436 pKa = 6.91HH437 pKa = 7.24IIVQIAAKK445 pKa = 8.89RR446 pKa = 11.84TGFQGLFTEE455 pKa = 4.55YY456 pKa = 10.75VILGDD461 pKa = 5.11DD462 pKa = 3.2ICIADD467 pKa = 4.55DD468 pKa = 4.45LVAAKK473 pKa = 9.65YY474 pKa = 10.3KK475 pKa = 11.09EE476 pKa = 3.88MMIEE480 pKa = 4.14LGLTISEE487 pKa = 4.52HH488 pKa = 5.04KK489 pKa = 9.66TIVSTVEE496 pKa = 3.65TSIDD500 pKa = 3.54PVAEE504 pKa = 3.41ICKK507 pKa = 9.69RR508 pKa = 11.84VFIGGVEE515 pKa = 4.09VTPLPMKK522 pKa = 10.45LAANVTEE529 pKa = 4.13NNDD532 pKa = 3.06LFFQFRR538 pKa = 11.84EE539 pKa = 4.16KK540 pKa = 10.72LSEE543 pKa = 4.11RR544 pKa = 11.84GLIYY548 pKa = 10.97NPADD552 pKa = 2.84WKK554 pKa = 11.05YY555 pKa = 10.29FVAACVSNHH564 pKa = 6.15RR565 pKa = 11.84SLEE568 pKa = 4.11KK569 pKa = 10.16IGLYY573 pKa = 10.76NSMPLWLSGFKK584 pKa = 10.16QSLPIGTVGNNNFEE598 pKa = 4.53DD599 pKa = 3.63WLKK602 pKa = 11.45AGMTEE607 pKa = 4.31DD608 pKa = 3.71YY609 pKa = 10.79LRR611 pKa = 11.84EE612 pKa = 3.84VWTFTVLQEE621 pKa = 3.84QFKK624 pKa = 10.71RR625 pKa = 11.84ISTLIQSAQDD635 pKa = 3.38TFQTISKK642 pKa = 9.8GIRR645 pKa = 11.84LGDD648 pKa = 3.41AVYY651 pKa = 10.1IVPDD655 pKa = 3.55TGIEE659 pKa = 4.09LPVTKK664 pKa = 10.31FDD666 pKa = 3.5IQKK669 pKa = 8.83MNEE672 pKa = 3.69WNILNIPHH680 pKa = 7.67PARR683 pKa = 11.84FVMLGEE689 pKa = 4.25VNRR692 pKa = 11.84VATLFSQLSSIRR704 pKa = 11.84GPSLMTKK711 pKa = 8.85LTHH714 pKa = 7.19NIVDD718 pKa = 3.59NLKK721 pKa = 10.45FSVLEE726 pKa = 4.62MINDD730 pKa = 3.32MDD732 pKa = 4.05NYY734 pKa = 9.42KK735 pKa = 10.89ARR737 pKa = 11.84VDD739 pKa = 3.65RR740 pKa = 11.84QLLEE744 pKa = 4.33KK745 pKa = 9.62MISNIRR751 pKa = 11.84MIIRR755 pKa = 11.84AEE757 pKa = 4.1NKK759 pKa = 8.45TVTYY763 pKa = 8.42TVKK766 pKa = 9.76LTPLGIVWVLRR777 pKa = 11.84LSHH780 pKa = 6.98LGSCTLFRR788 pKa = 11.84STSAIPTTLRR798 pKa = 11.84DD799 pKa = 3.44AEE801 pKa = 4.41LKK803 pKa = 9.79FKK805 pKa = 10.88RR806 pKa = 11.84MNSGNSQIASIFDD819 pKa = 3.48TT820 pKa = 4.34
Molecular weight: 92.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
820 |
820 |
820 |
820.0 |
92.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.854 ± 0.0 | 0.976 ± 0.0 |
5.122 ± 0.0 | 4.756 ± 0.0 |
4.39 ± 0.0 | 4.512 ± 0.0 |
1.829 ± 0.0 | 7.927 ± 0.0 |
6.707 ± 0.0 | 10.732 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.683 ± 0.0 | 6.098 ± 0.0 |
3.659 ± 0.0 | 4.268 ± 0.0 |
4.512 ± 0.0 | 7.805 ± 0.0 |
7.805 ± 0.0 | 6.341 ± 0.0 |
1.585 ± 0.0 | 2.439 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
