
Hubei sobemo-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
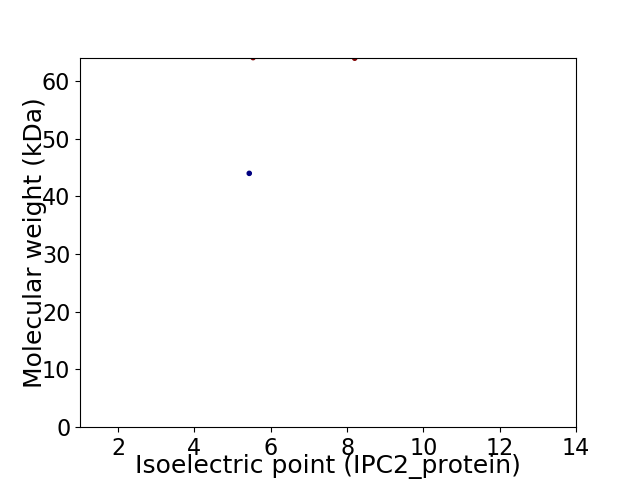
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEY4|A0A1L3KEY4_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 15 OX=1923200 PE=4 SV=1
MM1 pKa = 7.26VSASTGRR8 pKa = 11.84VEE10 pKa = 5.55FSVDD14 pKa = 3.12TQRR17 pKa = 11.84QLFDD21 pKa = 3.93AALEE25 pKa = 4.18DD26 pKa = 4.47LNMTSSVGKK35 pKa = 9.79CQMDD39 pKa = 3.66RR40 pKa = 11.84YY41 pKa = 10.08GATIGVALGYY51 pKa = 10.84DD52 pKa = 4.37PIAGCNQEE60 pKa = 5.06RR61 pKa = 11.84VDD63 pKa = 4.55FLYY66 pKa = 10.96HH67 pKa = 6.13LVSEE71 pKa = 4.78RR72 pKa = 11.84LEE74 pKa = 4.01EE75 pKa = 4.01MRR77 pKa = 11.84GGRR80 pKa = 11.84LVSDD84 pKa = 5.2PINLFVKK91 pKa = 10.17QEE93 pKa = 3.75PHH95 pKa = 5.77KK96 pKa = 10.2QSKK99 pKa = 9.2IRR101 pKa = 11.84EE102 pKa = 3.88GRR104 pKa = 11.84YY105 pKa = 9.33RR106 pKa = 11.84LISAVSLVDD115 pKa = 3.17TMIDD119 pKa = 3.12RR120 pKa = 11.84MIFGHH125 pKa = 6.64IKK127 pKa = 9.22QDD129 pKa = 2.93AWQDD133 pKa = 3.71SPVLIGWTPRR143 pKa = 11.84QGGYY147 pKa = 8.74RR148 pKa = 11.84WLWEE152 pKa = 3.88RR153 pKa = 11.84LDD155 pKa = 3.55GGKK158 pKa = 9.85RR159 pKa = 11.84LEE161 pKa = 3.94IDD163 pKa = 2.76KK164 pKa = 10.92RR165 pKa = 11.84AWDD168 pKa = 3.47WSMQPWVVRR177 pKa = 11.84GCLEE181 pKa = 3.86VLEE184 pKa = 4.33QLSLHH189 pKa = 6.35HH190 pKa = 6.32TDD192 pKa = 2.94EE193 pKa = 4.21WLKK196 pKa = 10.27LARR199 pKa = 11.84SRR201 pKa = 11.84FEE203 pKa = 3.71ALFYY207 pKa = 10.58RR208 pKa = 11.84ARR210 pKa = 11.84FDD212 pKa = 3.89GAGLTFDD219 pKa = 3.41QAEE222 pKa = 4.09PGIMKK227 pKa = 10.04SGCFLTIFANSLSQLLLHH245 pKa = 6.28FVAVRR250 pKa = 11.84RR251 pKa = 11.84SRR253 pKa = 11.84LGNLIEE259 pKa = 5.57APFCMGDD266 pKa = 3.84DD267 pKa = 4.38TIQRR271 pKa = 11.84CWASEE276 pKa = 4.1DD277 pKa = 3.61QVDD280 pKa = 3.99EE281 pKa = 4.18YY282 pKa = 11.65VRR284 pKa = 11.84QLEE287 pKa = 4.15EE288 pKa = 4.12AGCVVGDD295 pKa = 4.91CEE297 pKa = 4.84LLPAGSDD304 pKa = 3.57FQFCGFRR311 pKa = 11.84ITKK314 pKa = 10.06NKK316 pKa = 9.19VVPAYY321 pKa = 7.31TAKK324 pKa = 10.35HH325 pKa = 5.75CFVLQHH331 pKa = 6.72LEE333 pKa = 3.97DD334 pKa = 4.05DD335 pKa = 4.37TAVEE339 pKa = 4.41TLDD342 pKa = 5.5SYY344 pKa = 9.71QQDD347 pKa = 4.2YY348 pKa = 10.97VFEE351 pKa = 3.96PHH353 pKa = 6.85IYY355 pKa = 10.34GHH357 pKa = 6.57IADD360 pKa = 4.81GLLEE364 pKa = 4.08IRR366 pKa = 11.84PNALRR371 pKa = 11.84SKK373 pKa = 10.38GKK375 pKa = 8.13MRR377 pKa = 11.84RR378 pKa = 11.84FLRR381 pKa = 11.84GDD383 pKa = 3.31EE384 pKa = 4.08
MM1 pKa = 7.26VSASTGRR8 pKa = 11.84VEE10 pKa = 5.55FSVDD14 pKa = 3.12TQRR17 pKa = 11.84QLFDD21 pKa = 3.93AALEE25 pKa = 4.18DD26 pKa = 4.47LNMTSSVGKK35 pKa = 9.79CQMDD39 pKa = 3.66RR40 pKa = 11.84YY41 pKa = 10.08GATIGVALGYY51 pKa = 10.84DD52 pKa = 4.37PIAGCNQEE60 pKa = 5.06RR61 pKa = 11.84VDD63 pKa = 4.55FLYY66 pKa = 10.96HH67 pKa = 6.13LVSEE71 pKa = 4.78RR72 pKa = 11.84LEE74 pKa = 4.01EE75 pKa = 4.01MRR77 pKa = 11.84GGRR80 pKa = 11.84LVSDD84 pKa = 5.2PINLFVKK91 pKa = 10.17QEE93 pKa = 3.75PHH95 pKa = 5.77KK96 pKa = 10.2QSKK99 pKa = 9.2IRR101 pKa = 11.84EE102 pKa = 3.88GRR104 pKa = 11.84YY105 pKa = 9.33RR106 pKa = 11.84LISAVSLVDD115 pKa = 3.17TMIDD119 pKa = 3.12RR120 pKa = 11.84MIFGHH125 pKa = 6.64IKK127 pKa = 9.22QDD129 pKa = 2.93AWQDD133 pKa = 3.71SPVLIGWTPRR143 pKa = 11.84QGGYY147 pKa = 8.74RR148 pKa = 11.84WLWEE152 pKa = 3.88RR153 pKa = 11.84LDD155 pKa = 3.55GGKK158 pKa = 9.85RR159 pKa = 11.84LEE161 pKa = 3.94IDD163 pKa = 2.76KK164 pKa = 10.92RR165 pKa = 11.84AWDD168 pKa = 3.47WSMQPWVVRR177 pKa = 11.84GCLEE181 pKa = 3.86VLEE184 pKa = 4.33QLSLHH189 pKa = 6.35HH190 pKa = 6.32TDD192 pKa = 2.94EE193 pKa = 4.21WLKK196 pKa = 10.27LARR199 pKa = 11.84SRR201 pKa = 11.84FEE203 pKa = 3.71ALFYY207 pKa = 10.58RR208 pKa = 11.84ARR210 pKa = 11.84FDD212 pKa = 3.89GAGLTFDD219 pKa = 3.41QAEE222 pKa = 4.09PGIMKK227 pKa = 10.04SGCFLTIFANSLSQLLLHH245 pKa = 6.28FVAVRR250 pKa = 11.84RR251 pKa = 11.84SRR253 pKa = 11.84LGNLIEE259 pKa = 5.57APFCMGDD266 pKa = 3.84DD267 pKa = 4.38TIQRR271 pKa = 11.84CWASEE276 pKa = 4.1DD277 pKa = 3.61QVDD280 pKa = 3.99EE281 pKa = 4.18YY282 pKa = 11.65VRR284 pKa = 11.84QLEE287 pKa = 4.15EE288 pKa = 4.12AGCVVGDD295 pKa = 4.91CEE297 pKa = 4.84LLPAGSDD304 pKa = 3.57FQFCGFRR311 pKa = 11.84ITKK314 pKa = 10.06NKK316 pKa = 9.19VVPAYY321 pKa = 7.31TAKK324 pKa = 10.35HH325 pKa = 5.75CFVLQHH331 pKa = 6.72LEE333 pKa = 3.97DD334 pKa = 4.05DD335 pKa = 4.37TAVEE339 pKa = 4.41TLDD342 pKa = 5.5SYY344 pKa = 9.71QQDD347 pKa = 4.2YY348 pKa = 10.97VFEE351 pKa = 3.96PHH353 pKa = 6.85IYY355 pKa = 10.34GHH357 pKa = 6.57IADD360 pKa = 4.81GLLEE364 pKa = 4.08IRR366 pKa = 11.84PNALRR371 pKa = 11.84SKK373 pKa = 10.38GKK375 pKa = 8.13MRR377 pKa = 11.84RR378 pKa = 11.84FLRR381 pKa = 11.84GDD383 pKa = 3.31EE384 pKa = 4.08
Molecular weight: 43.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEY4|A0A1L3KEY4_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 15 OX=1923200 PE=4 SV=1
MM1 pKa = 7.21LQAGSQRR8 pKa = 11.84EE9 pKa = 4.26HH10 pKa = 6.9VLLAVLIAVTVGFFWLDD27 pKa = 2.75VHH29 pKa = 6.18RR30 pKa = 11.84QGVRR34 pKa = 11.84AEE36 pKa = 4.26LVSEE40 pKa = 4.27SMKK43 pKa = 10.9DD44 pKa = 3.38LNTLRR49 pKa = 11.84RR50 pKa = 11.84ILEE53 pKa = 4.06LNQRR57 pKa = 11.84LLRR60 pKa = 11.84NCQSTVFHH68 pKa = 6.97LKK70 pKa = 8.89QEE72 pKa = 4.19RR73 pKa = 11.84AQMDD77 pKa = 3.72RR78 pKa = 11.84VVARR82 pKa = 11.84LAAARR87 pKa = 11.84EE88 pKa = 4.44EE89 pKa = 4.41ACSPRR94 pKa = 11.84AADD97 pKa = 3.74RR98 pKa = 11.84VVSAAGDD105 pKa = 3.53AASWVAGAAAFGFALAWSAVAAYY128 pKa = 6.34WTLAFGVVGVVCMSYY143 pKa = 10.59VLWRR147 pKa = 11.84LLCLVYY153 pKa = 10.1RR154 pKa = 11.84WVRR157 pKa = 11.84EE158 pKa = 4.19RR159 pKa = 11.84IWNWRR164 pKa = 11.84APMVVEE170 pKa = 4.25EE171 pKa = 4.55QPLDD175 pKa = 3.98GPTLGCATLQWPGVEE190 pKa = 4.17IQPEE194 pKa = 4.79RR195 pKa = 11.84YY196 pKa = 8.81QAGSDD201 pKa = 3.56PHH203 pKa = 6.55PVSLKK208 pKa = 10.39DD209 pKa = 3.61LPPFQVAVYY218 pKa = 8.79TKK220 pKa = 10.62GAGKK224 pKa = 8.49TEE226 pKa = 4.53TYY228 pKa = 9.67FQGYY232 pKa = 9.11ALNIEE237 pKa = 5.17GYY239 pKa = 10.7LVMPLHH245 pKa = 5.99VKK247 pKa = 10.03EE248 pKa = 4.56PEE250 pKa = 4.46GVPHH254 pKa = 7.07DD255 pKa = 3.9HH256 pKa = 7.17LVLQRR261 pKa = 11.84YY262 pKa = 9.04VGDD265 pKa = 3.86EE266 pKa = 3.92LRR268 pKa = 11.84QTTLGEE274 pKa = 4.26TITWMEE280 pKa = 4.32LAQDD284 pKa = 3.78AVAADD289 pKa = 3.89MSTMKK294 pKa = 10.13FDD296 pKa = 3.59KK297 pKa = 11.19GNLEE301 pKa = 4.28SYY303 pKa = 10.67LGLAKK308 pKa = 10.62AKK310 pKa = 10.24VADD313 pKa = 3.94VTEE316 pKa = 4.37MTHH319 pKa = 6.45CSIVGGFRR327 pKa = 11.84PVARR331 pKa = 11.84TMGAVRR337 pKa = 11.84LLSEE341 pKa = 4.58FFGQVQYY348 pKa = 11.11TGTTFPGYY356 pKa = 10.15SGAAYY361 pKa = 9.62YY362 pKa = 10.51VNKK365 pKa = 9.1TVYY368 pKa = 10.7GMHH371 pKa = 6.24TSAWHH376 pKa = 5.11TMNVGYY382 pKa = 10.02SAAYY386 pKa = 9.04LASILRR392 pKa = 11.84KK393 pKa = 9.6RR394 pKa = 11.84RR395 pKa = 11.84GQKK398 pKa = 9.75EE399 pKa = 3.99SSEE402 pKa = 4.43DD403 pKa = 3.49FLFQRR408 pKa = 11.84MTKK411 pKa = 8.8QKK413 pKa = 10.3KK414 pKa = 8.3ILYY417 pKa = 7.02RR418 pKa = 11.84TSPGDD423 pKa = 3.49PEE425 pKa = 4.06EE426 pKa = 5.51VIMKK430 pKa = 9.99VEE432 pKa = 3.8GRR434 pKa = 11.84FYY436 pKa = 10.71TFARR440 pKa = 11.84DD441 pKa = 3.59EE442 pKa = 4.33IPRR445 pKa = 11.84SVWDD449 pKa = 3.84RR450 pKa = 11.84MVSADD455 pKa = 3.82DD456 pKa = 3.72VQGSGEE462 pKa = 4.29SYY464 pKa = 10.84NDD466 pKa = 3.14EE467 pKa = 4.09GCFLGRR473 pKa = 11.84RR474 pKa = 11.84PAQSQSLGQMGQLKK488 pKa = 10.25RR489 pKa = 11.84MPDD492 pKa = 3.17NRR494 pKa = 11.84SPGVSVSQHH503 pKa = 4.53GQQTIPNVAAGLSGQTPEE521 pKa = 4.18RR522 pKa = 11.84TSGSSSAKK530 pKa = 9.17NAEE533 pKa = 4.15GKK535 pKa = 9.21PEE537 pKa = 3.87NMVSQEE543 pKa = 4.2RR544 pKa = 11.84IVVQSPPPSSGTSSITEE561 pKa = 3.72QSATGSKK568 pKa = 9.77SRR570 pKa = 11.84RR571 pKa = 11.84NKK573 pKa = 10.12SSKK576 pKa = 10.54KK577 pKa = 9.12PP578 pKa = 3.44
MM1 pKa = 7.21LQAGSQRR8 pKa = 11.84EE9 pKa = 4.26HH10 pKa = 6.9VLLAVLIAVTVGFFWLDD27 pKa = 2.75VHH29 pKa = 6.18RR30 pKa = 11.84QGVRR34 pKa = 11.84AEE36 pKa = 4.26LVSEE40 pKa = 4.27SMKK43 pKa = 10.9DD44 pKa = 3.38LNTLRR49 pKa = 11.84RR50 pKa = 11.84ILEE53 pKa = 4.06LNQRR57 pKa = 11.84LLRR60 pKa = 11.84NCQSTVFHH68 pKa = 6.97LKK70 pKa = 8.89QEE72 pKa = 4.19RR73 pKa = 11.84AQMDD77 pKa = 3.72RR78 pKa = 11.84VVARR82 pKa = 11.84LAAARR87 pKa = 11.84EE88 pKa = 4.44EE89 pKa = 4.41ACSPRR94 pKa = 11.84AADD97 pKa = 3.74RR98 pKa = 11.84VVSAAGDD105 pKa = 3.53AASWVAGAAAFGFALAWSAVAAYY128 pKa = 6.34WTLAFGVVGVVCMSYY143 pKa = 10.59VLWRR147 pKa = 11.84LLCLVYY153 pKa = 10.1RR154 pKa = 11.84WVRR157 pKa = 11.84EE158 pKa = 4.19RR159 pKa = 11.84IWNWRR164 pKa = 11.84APMVVEE170 pKa = 4.25EE171 pKa = 4.55QPLDD175 pKa = 3.98GPTLGCATLQWPGVEE190 pKa = 4.17IQPEE194 pKa = 4.79RR195 pKa = 11.84YY196 pKa = 8.81QAGSDD201 pKa = 3.56PHH203 pKa = 6.55PVSLKK208 pKa = 10.39DD209 pKa = 3.61LPPFQVAVYY218 pKa = 8.79TKK220 pKa = 10.62GAGKK224 pKa = 8.49TEE226 pKa = 4.53TYY228 pKa = 9.67FQGYY232 pKa = 9.11ALNIEE237 pKa = 5.17GYY239 pKa = 10.7LVMPLHH245 pKa = 5.99VKK247 pKa = 10.03EE248 pKa = 4.56PEE250 pKa = 4.46GVPHH254 pKa = 7.07DD255 pKa = 3.9HH256 pKa = 7.17LVLQRR261 pKa = 11.84YY262 pKa = 9.04VGDD265 pKa = 3.86EE266 pKa = 3.92LRR268 pKa = 11.84QTTLGEE274 pKa = 4.26TITWMEE280 pKa = 4.32LAQDD284 pKa = 3.78AVAADD289 pKa = 3.89MSTMKK294 pKa = 10.13FDD296 pKa = 3.59KK297 pKa = 11.19GNLEE301 pKa = 4.28SYY303 pKa = 10.67LGLAKK308 pKa = 10.62AKK310 pKa = 10.24VADD313 pKa = 3.94VTEE316 pKa = 4.37MTHH319 pKa = 6.45CSIVGGFRR327 pKa = 11.84PVARR331 pKa = 11.84TMGAVRR337 pKa = 11.84LLSEE341 pKa = 4.58FFGQVQYY348 pKa = 11.11TGTTFPGYY356 pKa = 10.15SGAAYY361 pKa = 9.62YY362 pKa = 10.51VNKK365 pKa = 9.1TVYY368 pKa = 10.7GMHH371 pKa = 6.24TSAWHH376 pKa = 5.11TMNVGYY382 pKa = 10.02SAAYY386 pKa = 9.04LASILRR392 pKa = 11.84KK393 pKa = 9.6RR394 pKa = 11.84RR395 pKa = 11.84GQKK398 pKa = 9.75EE399 pKa = 3.99SSEE402 pKa = 4.43DD403 pKa = 3.49FLFQRR408 pKa = 11.84MTKK411 pKa = 8.8QKK413 pKa = 10.3KK414 pKa = 8.3ILYY417 pKa = 7.02RR418 pKa = 11.84TSPGDD423 pKa = 3.49PEE425 pKa = 4.06EE426 pKa = 5.51VIMKK430 pKa = 9.99VEE432 pKa = 3.8GRR434 pKa = 11.84FYY436 pKa = 10.71TFARR440 pKa = 11.84DD441 pKa = 3.59EE442 pKa = 4.33IPRR445 pKa = 11.84SVWDD449 pKa = 3.84RR450 pKa = 11.84MVSADD455 pKa = 3.82DD456 pKa = 3.72VQGSGEE462 pKa = 4.29SYY464 pKa = 10.84NDD466 pKa = 3.14EE467 pKa = 4.09GCFLGRR473 pKa = 11.84RR474 pKa = 11.84PAQSQSLGQMGQLKK488 pKa = 10.25RR489 pKa = 11.84MPDD492 pKa = 3.17NRR494 pKa = 11.84SPGVSVSQHH503 pKa = 4.53GQQTIPNVAAGLSGQTPEE521 pKa = 4.18RR522 pKa = 11.84TSGSSSAKK530 pKa = 9.17NAEE533 pKa = 4.15GKK535 pKa = 9.21PEE537 pKa = 3.87NMVSQEE543 pKa = 4.2RR544 pKa = 11.84IVVQSPPPSSGTSSITEE561 pKa = 3.72QSATGSKK568 pKa = 9.77SRR570 pKa = 11.84RR571 pKa = 11.84NKK573 pKa = 10.12SSKK576 pKa = 10.54KK577 pKa = 9.12PP578 pKa = 3.44
Molecular weight: 63.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
962 |
384 |
578 |
481.0 |
53.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.316 ± 1.131 | 1.767 ± 0.524 |
5.301 ± 1.41 | 6.341 ± 0.269 |
3.846 ± 0.69 | 8.004 ± 0.12 |
2.183 ± 0.264 | 3.326 ± 0.853 |
3.95 ± 0.191 | 8.94 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.015 ± 0.257 | 2.183 ± 0.226 |
4.158 ± 0.647 | 5.509 ± 0.189 |
7.588 ± 0.467 | 7.173 ± 0.904 |
4.782 ± 0.712 | 8.212 ± 0.903 |
2.183 ± 0.101 | 3.222 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
