
uncultured phage_MedDCM-OCT-S46-C10
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Foussvirus; Foussvirus S46C10
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
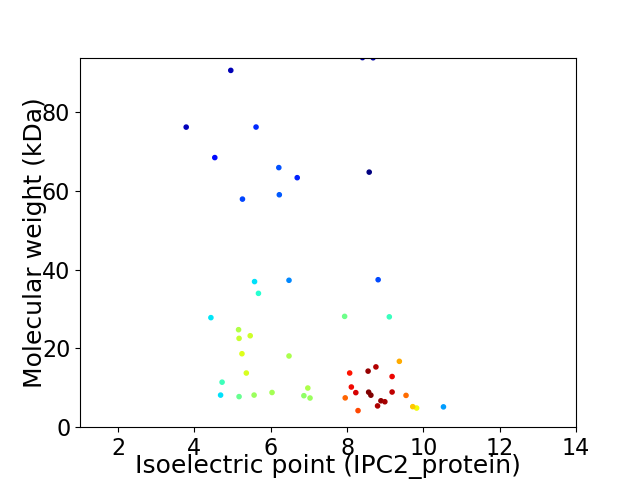
Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6S4PDZ4|A0A6S4PDZ4_9CAUD Uncharacterized protein OS=uncultured phage_MedDCM-OCT-S46-C10 OX=2741074 PE=4 SV=1
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.35TGNGNTTAYY17 pKa = 10.02SIPYY21 pKa = 9.41SYY23 pKa = 10.68RR24 pKa = 11.84DD25 pKa = 3.51PADD28 pKa = 4.47LIVTINGVATTSYY41 pKa = 9.57TLNSAGSTLTFDD53 pKa = 3.73TAPANSSAIEE63 pKa = 3.91IRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.21TSQTSRR73 pKa = 11.84LTDD76 pKa = 3.47YY77 pKa = 11.3AAGSVLTEE85 pKa = 4.12NDD87 pKa = 3.84LDD89 pKa = 4.08TDD91 pKa = 3.79STQAFFMSQEE101 pKa = 4.56AIDD104 pKa = 4.32DD105 pKa = 4.19AGDD108 pKa = 3.7VIKK111 pKa = 10.95LSNTNFQWDD120 pKa = 4.08TQNKK124 pKa = 9.3RR125 pKa = 11.84LTNVADD131 pKa = 3.91PVNNTDD137 pKa = 3.4GVNKK141 pKa = 9.92QFISTNLPNITTVSGISSDD160 pKa = 3.52VTTVAGIASNVTAVASDD177 pKa = 3.27ATDD180 pKa = 2.91IGTVATNIASVNTVATDD197 pKa = 3.25IAKK200 pKa = 10.57VIVVANDD207 pKa = 3.31LNEE210 pKa = 4.25TVSEE214 pKa = 4.29IEE216 pKa = 3.98TAALDD221 pKa = 3.94LQEE224 pKa = 4.36TTSEE228 pKa = 3.89IDD230 pKa = 3.42TVSNNIANVNTVGTNITNVNTVAGVSANVTTVAGINTDD268 pKa = 3.39VTSVAGISSAVSAVNSNSTNINAVNANSANINTVAGIDD306 pKa = 3.58SDD308 pKa = 3.66ITSVANISSDD318 pKa = 3.3VAAVEE323 pKa = 4.42NIAANVTTVAGNNANITTVAGANSNITAVAGAITNVNNVGGAIANVNNVGGSIANVNTVATNLASVNNFAEE394 pKa = 4.46QYY396 pKa = 9.61RR397 pKa = 11.84IASSAPTSSLNVGDD411 pKa = 5.36LYY413 pKa = 11.47FDD415 pKa = 3.51TTANEE420 pKa = 3.92LKK422 pKa = 10.5VYY424 pKa = 10.44KK425 pKa = 10.46SSGWAAAGSTVNGTSQRR442 pKa = 11.84FTYY445 pKa = 9.36TISGTPSSVTGSDD458 pKa = 3.39ANGNTLAYY466 pKa = 9.92DD467 pKa = 3.42AGFADD472 pKa = 4.41VYY474 pKa = 11.19LNGVRR479 pKa = 11.84LSSADD484 pKa = 3.13ITITSGTSVVFASALANGDD503 pKa = 3.63VVDD506 pKa = 4.41VVAYY510 pKa = 7.85GTFNVASINAANISSGTVNNARR532 pKa = 11.84LTGSGAITINGSSVSLGGNITVGEE556 pKa = 4.34TKK558 pKa = 9.09PTIGSISPSTITNAQTSITITGTNFVSVPQVEE590 pKa = 4.8ALNQSTGIWYY600 pKa = 7.87TADD603 pKa = 3.45TISFTNATTLVATFTLSVDD622 pKa = 3.09AQYY625 pKa = 11.81KK626 pKa = 10.17LRR628 pKa = 11.84IEE630 pKa = 4.49NPDD633 pKa = 3.08GNAVLSSSNILTVSDD648 pKa = 3.74APTWSTSAGSLGVFAGNFSGTLATISASSDD678 pKa = 3.18STVAYY683 pKa = 10.72SEE685 pKa = 4.44TTSVLTGAGVTLNTSTGALTTSDD708 pKa = 4.38FGASSTTPTTYY719 pKa = 11.01NFTIRR724 pKa = 11.84ATDD727 pKa = 3.85GEE729 pKa = 4.7GQTTDD734 pKa = 3.29RR735 pKa = 11.84SFSMTSSFGSTGGGQFNN752 pKa = 4.02
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.35TGNGNTTAYY17 pKa = 10.02SIPYY21 pKa = 9.41SYY23 pKa = 10.68RR24 pKa = 11.84DD25 pKa = 3.51PADD28 pKa = 4.47LIVTINGVATTSYY41 pKa = 9.57TLNSAGSTLTFDD53 pKa = 3.73TAPANSSAIEE63 pKa = 3.91IRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.21TSQTSRR73 pKa = 11.84LTDD76 pKa = 3.47YY77 pKa = 11.3AAGSVLTEE85 pKa = 4.12NDD87 pKa = 3.84LDD89 pKa = 4.08TDD91 pKa = 3.79STQAFFMSQEE101 pKa = 4.56AIDD104 pKa = 4.32DD105 pKa = 4.19AGDD108 pKa = 3.7VIKK111 pKa = 10.95LSNTNFQWDD120 pKa = 4.08TQNKK124 pKa = 9.3RR125 pKa = 11.84LTNVADD131 pKa = 3.91PVNNTDD137 pKa = 3.4GVNKK141 pKa = 9.92QFISTNLPNITTVSGISSDD160 pKa = 3.52VTTVAGIASNVTAVASDD177 pKa = 3.27ATDD180 pKa = 2.91IGTVATNIASVNTVATDD197 pKa = 3.25IAKK200 pKa = 10.57VIVVANDD207 pKa = 3.31LNEE210 pKa = 4.25TVSEE214 pKa = 4.29IEE216 pKa = 3.98TAALDD221 pKa = 3.94LQEE224 pKa = 4.36TTSEE228 pKa = 3.89IDD230 pKa = 3.42TVSNNIANVNTVGTNITNVNTVAGVSANVTTVAGINTDD268 pKa = 3.39VTSVAGISSAVSAVNSNSTNINAVNANSANINTVAGIDD306 pKa = 3.58SDD308 pKa = 3.66ITSVANISSDD318 pKa = 3.3VAAVEE323 pKa = 4.42NIAANVTTVAGNNANITTVAGANSNITAVAGAITNVNNVGGAIANVNNVGGSIANVNTVATNLASVNNFAEE394 pKa = 4.46QYY396 pKa = 9.61RR397 pKa = 11.84IASSAPTSSLNVGDD411 pKa = 5.36LYY413 pKa = 11.47FDD415 pKa = 3.51TTANEE420 pKa = 3.92LKK422 pKa = 10.5VYY424 pKa = 10.44KK425 pKa = 10.46SSGWAAAGSTVNGTSQRR442 pKa = 11.84FTYY445 pKa = 9.36TISGTPSSVTGSDD458 pKa = 3.39ANGNTLAYY466 pKa = 9.92DD467 pKa = 3.42AGFADD472 pKa = 4.41VYY474 pKa = 11.19LNGVRR479 pKa = 11.84LSSADD484 pKa = 3.13ITITSGTSVVFASALANGDD503 pKa = 3.63VVDD506 pKa = 4.41VVAYY510 pKa = 7.85GTFNVASINAANISSGTVNNARR532 pKa = 11.84LTGSGAITINGSSVSLGGNITVGEE556 pKa = 4.34TKK558 pKa = 9.09PTIGSISPSTITNAQTSITITGTNFVSVPQVEE590 pKa = 4.8ALNQSTGIWYY600 pKa = 7.87TADD603 pKa = 3.45TISFTNATTLVATFTLSVDD622 pKa = 3.09AQYY625 pKa = 11.81KK626 pKa = 10.17LRR628 pKa = 11.84IEE630 pKa = 4.49NPDD633 pKa = 3.08GNAVLSSSNILTVSDD648 pKa = 3.74APTWSTSAGSLGVFAGNFSGTLATISASSDD678 pKa = 3.18STVAYY683 pKa = 10.72SEE685 pKa = 4.44TTSVLTGAGVTLNTSTGALTTSDD708 pKa = 4.38FGASSTTPTTYY719 pKa = 11.01NFTIRR724 pKa = 11.84ATDD727 pKa = 3.85GEE729 pKa = 4.7GQTTDD734 pKa = 3.29RR735 pKa = 11.84SFSMTSSFGSTGGGQFNN752 pKa = 4.02
Molecular weight: 76.13 kDa
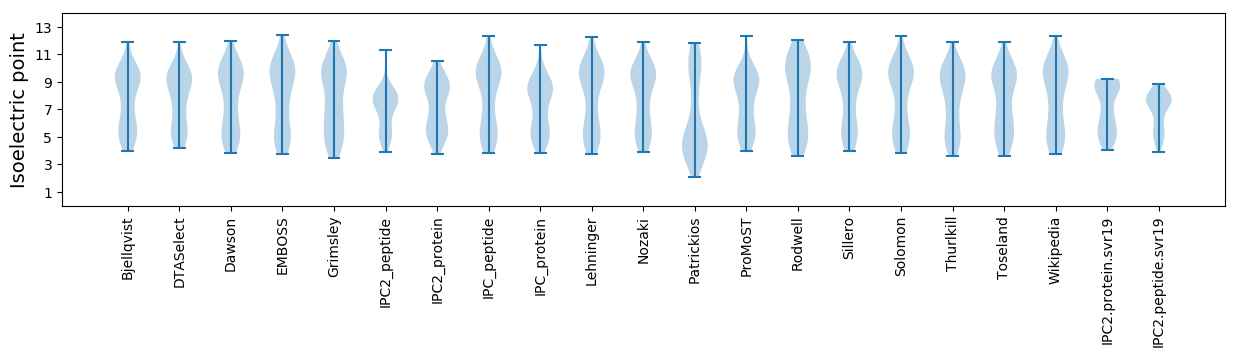
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6S4PIL5|A0A6S4PIL5_9CAUD Uncharacterized protein OS=uncultured phage_MedDCM-OCT-S46-C10 OX=2741074 PE=4 SV=1
MM1 pKa = 7.35YY2 pKa = 10.03MNFILFKK9 pKa = 9.81VTIEE13 pKa = 3.97RR14 pKa = 11.84WSKK17 pKa = 10.33RR18 pKa = 11.84SNLKK22 pKa = 7.48VTKK25 pKa = 10.32RR26 pKa = 11.84NGEE29 pKa = 4.27TIVDD33 pKa = 3.45VAYY36 pKa = 9.78WRR38 pKa = 11.84IYY40 pKa = 9.57LQQ42 pKa = 3.45
MM1 pKa = 7.35YY2 pKa = 10.03MNFILFKK9 pKa = 9.81VTIEE13 pKa = 3.97RR14 pKa = 11.84WSKK17 pKa = 10.33RR18 pKa = 11.84SNLKK22 pKa = 7.48VTKK25 pKa = 10.32RR26 pKa = 11.84NGEE29 pKa = 4.27TIVDD33 pKa = 3.45VAYY36 pKa = 9.78WRR38 pKa = 11.84IYY40 pKa = 9.57LQQ42 pKa = 3.45
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12493 |
38 |
832 |
245.0 |
27.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.148 ± 0.523 | 0.985 ± 0.175 |
5.771 ± 0.199 | 6.404 ± 0.443 |
3.882 ± 0.244 | 6.115 ± 0.296 |
1.673 ± 0.212 | 6.604 ± 0.232 |
8.333 ± 0.693 | 8.149 ± 0.425 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.497 ± 0.292 | 6.388 ± 0.449 |
2.962 ± 0.208 | 3.818 ± 0.26 |
4.09 ± 0.33 | 7.268 ± 0.459 |
7.244 ± 0.635 | 6.107 ± 0.357 |
1.257 ± 0.136 | 3.306 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
