
High Plains wheat mosaic emaravirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
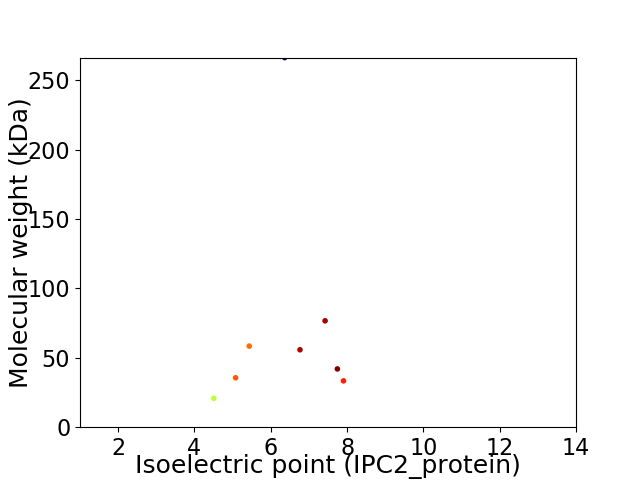
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A076VAA0|A0A076VAA0_9VIRU Uncharacterized protein OS=High Plains wheat mosaic emaravirus OX=1980428 PE=4 SV=1
MM1 pKa = 6.95NTIKK5 pKa = 9.95TVIISEE11 pKa = 4.26LEE13 pKa = 4.05KK14 pKa = 11.28NVDD17 pKa = 3.44EE18 pKa = 4.81FLNSYY23 pKa = 10.75LEE25 pKa = 3.89YY26 pKa = 10.78LKK28 pKa = 11.01YY29 pKa = 10.64DD30 pKa = 5.11DD31 pKa = 4.41YY32 pKa = 11.98DD33 pKa = 3.6QYY35 pKa = 11.53CTMIGLYY42 pKa = 10.45DD43 pKa = 3.99EE44 pKa = 5.57LTDD47 pKa = 4.29QEE49 pKa = 5.2SISQIPTKK57 pKa = 10.91YY58 pKa = 10.7SIDD61 pKa = 4.19PINFQKK67 pKa = 9.6FTRR70 pKa = 11.84VLTVAIYY77 pKa = 10.3NYY79 pKa = 10.2DD80 pKa = 3.18VNYY83 pKa = 10.37ILAEE87 pKa = 4.05KK88 pKa = 10.29YY89 pKa = 9.96KK90 pKa = 10.77EE91 pKa = 3.98LFEE94 pKa = 4.22FTNMDD99 pKa = 3.76PDD101 pKa = 4.35FSPKK105 pKa = 9.74YY106 pKa = 10.07RR107 pKa = 11.84FYY109 pKa = 11.59SPIATCSYY117 pKa = 10.28LSQYY121 pKa = 11.46DD122 pKa = 4.44LISEE126 pKa = 4.56SFQQDD131 pKa = 3.18VTKK134 pKa = 10.95LFDD137 pKa = 4.21RR138 pKa = 11.84MHH140 pKa = 6.21KK141 pKa = 9.04QQPGCMLMNQIMVSNLIKK159 pKa = 10.84NLLKK163 pKa = 10.55NVQTIGQDD171 pKa = 3.5DD172 pKa = 4.0SSHH175 pKa = 5.3NN176 pKa = 3.64
MM1 pKa = 6.95NTIKK5 pKa = 9.95TVIISEE11 pKa = 4.26LEE13 pKa = 4.05KK14 pKa = 11.28NVDD17 pKa = 3.44EE18 pKa = 4.81FLNSYY23 pKa = 10.75LEE25 pKa = 3.89YY26 pKa = 10.78LKK28 pKa = 11.01YY29 pKa = 10.64DD30 pKa = 5.11DD31 pKa = 4.41YY32 pKa = 11.98DD33 pKa = 3.6QYY35 pKa = 11.53CTMIGLYY42 pKa = 10.45DD43 pKa = 3.99EE44 pKa = 5.57LTDD47 pKa = 4.29QEE49 pKa = 5.2SISQIPTKK57 pKa = 10.91YY58 pKa = 10.7SIDD61 pKa = 4.19PINFQKK67 pKa = 9.6FTRR70 pKa = 11.84VLTVAIYY77 pKa = 10.3NYY79 pKa = 10.2DD80 pKa = 3.18VNYY83 pKa = 10.37ILAEE87 pKa = 4.05KK88 pKa = 10.29YY89 pKa = 9.96KK90 pKa = 10.77EE91 pKa = 3.98LFEE94 pKa = 4.22FTNMDD99 pKa = 3.76PDD101 pKa = 4.35FSPKK105 pKa = 9.74YY106 pKa = 10.07RR107 pKa = 11.84FYY109 pKa = 11.59SPIATCSYY117 pKa = 10.28LSQYY121 pKa = 11.46DD122 pKa = 4.44LISEE126 pKa = 4.56SFQQDD131 pKa = 3.18VTKK134 pKa = 10.95LFDD137 pKa = 4.21RR138 pKa = 11.84MHH140 pKa = 6.21KK141 pKa = 9.04QQPGCMLMNQIMVSNLIKK159 pKa = 10.84NLLKK163 pKa = 10.55NVQTIGQDD171 pKa = 3.5DD172 pKa = 4.0SSHH175 pKa = 5.3NN176 pKa = 3.64
Molecular weight: 20.82 kDa
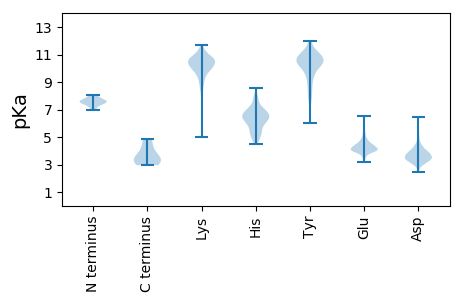
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076V8I4|A0A076V8I4_9VIRU P8 RNA silencing suppressor OS=High Plains wheat mosaic emaravirus OX=1980428 PE=4 SV=1
MM1 pKa = 7.55ALSFKK6 pKa = 10.64NSSGVIKK13 pKa = 10.73AKK15 pKa = 9.53TIKK18 pKa = 10.66DD19 pKa = 3.67GFVTSSDD26 pKa = 3.19IEE28 pKa = 4.55TTVHH32 pKa = 6.36DD33 pKa = 4.54FSYY36 pKa = 10.61EE37 pKa = 4.03KK38 pKa = 10.22PDD40 pKa = 3.64LSSVDD45 pKa = 3.59GFSLKK50 pKa = 10.72SLLSSDD56 pKa = 2.8GWHH59 pKa = 6.18IVVAYY64 pKa = 9.71QSVTNSEE71 pKa = 3.96RR72 pKa = 11.84LNNNKK77 pKa = 9.56KK78 pKa = 10.03NNKK81 pKa = 4.97TQRR84 pKa = 11.84FKK86 pKa = 11.55LFTFDD91 pKa = 3.99IIVIPGLKK99 pKa = 9.55PNKK102 pKa = 9.3SKK104 pKa = 11.29NVVSYY109 pKa = 10.84NRR111 pKa = 11.84FMALCIGMICYY122 pKa = 9.17HH123 pKa = 6.42KK124 pKa = 10.13KK125 pKa = 9.06WKK127 pKa = 9.24VFNWSNKK134 pKa = 9.0RR135 pKa = 11.84YY136 pKa = 9.46EE137 pKa = 4.32DD138 pKa = 3.44NKK140 pKa = 9.82NTINFNEE147 pKa = 5.23DD148 pKa = 3.16DD149 pKa = 4.3DD150 pKa = 4.96FMNKK154 pKa = 9.44LAMSAGFSKK163 pKa = 9.4EE164 pKa = 4.14HH165 pKa = 6.67KK166 pKa = 9.25YY167 pKa = 10.28HH168 pKa = 6.59WFYY171 pKa = 11.25STGFEE176 pKa = 4.17YY177 pKa = 10.72TFDD180 pKa = 3.43IFPAEE185 pKa = 4.39VIAMSLFRR193 pKa = 11.84WSHH196 pKa = 4.55RR197 pKa = 11.84VEE199 pKa = 4.7LKK201 pKa = 10.52VKK203 pKa = 10.36YY204 pKa = 8.33EE205 pKa = 4.08HH206 pKa = 7.03EE207 pKa = 4.63SDD209 pKa = 4.05LVAPMVRR216 pKa = 11.84QVTKK220 pKa = 10.22RR221 pKa = 11.84GNISDD226 pKa = 3.62VMDD229 pKa = 3.61IVGKK233 pKa = 10.24DD234 pKa = 3.43IIAKK238 pKa = 9.76KK239 pKa = 10.32YY240 pKa = 9.96EE241 pKa = 4.43EE242 pKa = 4.13IVKK245 pKa = 10.14DD246 pKa = 3.85RR247 pKa = 11.84SSIGIGTKK255 pKa = 10.68YY256 pKa = 10.99NDD258 pKa = 3.58ILDD261 pKa = 3.82EE262 pKa = 4.8FKK264 pKa = 11.11DD265 pKa = 3.48IFNKK269 pKa = 9.76IDD271 pKa = 3.57SSSLDD276 pKa = 3.53STIKK280 pKa = 10.79NCFNKK285 pKa = 9.66IDD287 pKa = 3.92GEE289 pKa = 4.34
MM1 pKa = 7.55ALSFKK6 pKa = 10.64NSSGVIKK13 pKa = 10.73AKK15 pKa = 9.53TIKK18 pKa = 10.66DD19 pKa = 3.67GFVTSSDD26 pKa = 3.19IEE28 pKa = 4.55TTVHH32 pKa = 6.36DD33 pKa = 4.54FSYY36 pKa = 10.61EE37 pKa = 4.03KK38 pKa = 10.22PDD40 pKa = 3.64LSSVDD45 pKa = 3.59GFSLKK50 pKa = 10.72SLLSSDD56 pKa = 2.8GWHH59 pKa = 6.18IVVAYY64 pKa = 9.71QSVTNSEE71 pKa = 3.96RR72 pKa = 11.84LNNNKK77 pKa = 9.56KK78 pKa = 10.03NNKK81 pKa = 4.97TQRR84 pKa = 11.84FKK86 pKa = 11.55LFTFDD91 pKa = 3.99IIVIPGLKK99 pKa = 9.55PNKK102 pKa = 9.3SKK104 pKa = 11.29NVVSYY109 pKa = 10.84NRR111 pKa = 11.84FMALCIGMICYY122 pKa = 9.17HH123 pKa = 6.42KK124 pKa = 10.13KK125 pKa = 9.06WKK127 pKa = 9.24VFNWSNKK134 pKa = 9.0RR135 pKa = 11.84YY136 pKa = 9.46EE137 pKa = 4.32DD138 pKa = 3.44NKK140 pKa = 9.82NTINFNEE147 pKa = 5.23DD148 pKa = 3.16DD149 pKa = 4.3DD150 pKa = 4.96FMNKK154 pKa = 9.44LAMSAGFSKK163 pKa = 9.4EE164 pKa = 4.14HH165 pKa = 6.67KK166 pKa = 9.25YY167 pKa = 10.28HH168 pKa = 6.59WFYY171 pKa = 11.25STGFEE176 pKa = 4.17YY177 pKa = 10.72TFDD180 pKa = 3.43IFPAEE185 pKa = 4.39VIAMSLFRR193 pKa = 11.84WSHH196 pKa = 4.55RR197 pKa = 11.84VEE199 pKa = 4.7LKK201 pKa = 10.52VKK203 pKa = 10.36YY204 pKa = 8.33EE205 pKa = 4.08HH206 pKa = 7.03EE207 pKa = 4.63SDD209 pKa = 4.05LVAPMVRR216 pKa = 11.84QVTKK220 pKa = 10.22RR221 pKa = 11.84GNISDD226 pKa = 3.62VMDD229 pKa = 3.61IVGKK233 pKa = 10.24DD234 pKa = 3.43IIAKK238 pKa = 9.76KK239 pKa = 10.32YY240 pKa = 9.96EE241 pKa = 4.43EE242 pKa = 4.13IVKK245 pKa = 10.14DD246 pKa = 3.85RR247 pKa = 11.84SSIGIGTKK255 pKa = 10.68YY256 pKa = 10.99NDD258 pKa = 3.58ILDD261 pKa = 3.82EE262 pKa = 4.8FKK264 pKa = 11.11DD265 pKa = 3.48IFNKK269 pKa = 9.76IDD271 pKa = 3.57SSSLDD276 pKa = 3.53STIKK280 pKa = 10.79NCFNKK285 pKa = 9.66IDD287 pKa = 3.92GEE289 pKa = 4.34
Molecular weight: 33.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5043 |
176 |
2272 |
630.4 |
73.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.637 ± 0.242 | 1.765 ± 0.325 |
7.079 ± 0.349 | 6.246 ± 0.451 |
5.354 ± 0.35 | 3.193 ± 0.411 |
2.181 ± 0.193 | 8.467 ± 0.282 |
9.478 ± 0.375 | 8.745 ± 0.77 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.875 ± 0.192 | 7.476 ± 0.26 |
2.677 ± 0.246 | 2.796 ± 0.416 |
3.411 ± 0.333 | 8.031 ± 0.434 |
5.513 ± 0.28 | 5.731 ± 0.531 |
0.555 ± 0.198 | 5.79 ± 0.434 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
