
Synechococcus phage S-CBS2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
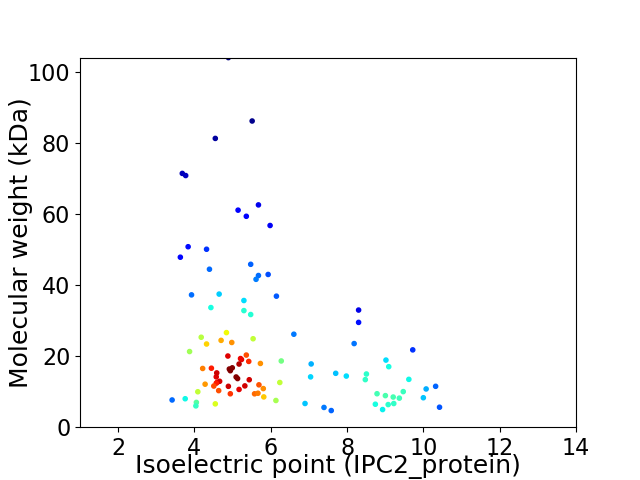
Virtual 2D-PAGE plot for 102 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
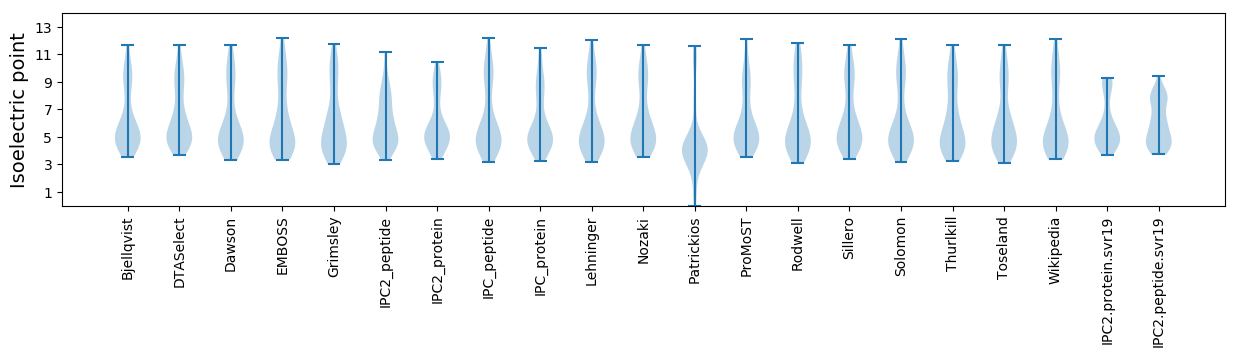
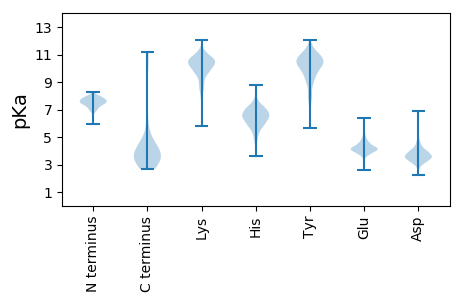
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4YCL3|F4YCL3_9CAUD Structural protein OS=Synechococcus phage S-CBS2 OX=753084 GN=S-CBS2_gp032 PE=4 SV=1
MM1 pKa = 7.63AVPSNRR7 pKa = 11.84VPVRR11 pKa = 11.84VARR14 pKa = 11.84GLKK17 pKa = 10.04SALTANLADD26 pKa = 4.16LLEE29 pKa = 5.37GEE31 pKa = 4.59LVYY34 pKa = 11.13AKK36 pKa = 10.76DD37 pKa = 3.53EE38 pKa = 4.7DD39 pKa = 3.97KK40 pKa = 11.33LYY42 pKa = 10.04MIEE45 pKa = 4.46GGTLVAMGADD55 pKa = 3.96LAAASILDD63 pKa = 4.14LGDD66 pKa = 3.71VEE68 pKa = 5.32PNSLPDD74 pKa = 3.44GARR77 pKa = 11.84YY78 pKa = 9.82ALGGVQFPGNGLWYY92 pKa = 10.07RR93 pKa = 11.84SGSRR97 pKa = 11.84ISWGAIDD104 pKa = 5.55ADD106 pKa = 3.96STDD109 pKa = 3.42RR110 pKa = 11.84TSQITAASGSIFWSTDD126 pKa = 2.43GATWNEE132 pKa = 3.73IFFTGSPQTVGDD144 pKa = 4.21SYY146 pKa = 11.7YY147 pKa = 10.24IDD149 pKa = 3.44ITSGSLPALTGDD161 pKa = 4.62LYY163 pKa = 11.35FSFSTPGTPSPAPLADD179 pKa = 3.49GDD181 pKa = 4.05YY182 pKa = 11.26LRR184 pKa = 11.84FDD186 pKa = 3.68GTDD189 pKa = 3.52FRR191 pKa = 11.84PQQLATVAEE200 pKa = 4.39TGAYY204 pKa = 9.7SDD206 pKa = 5.03LSGTPTLVEE215 pKa = 4.8NIDD218 pKa = 4.18DD219 pKa = 4.12LTDD222 pKa = 3.25VDD224 pKa = 4.57TSTSAPTTDD233 pKa = 5.61QVLAWNGTNWVPSDD247 pKa = 3.42VSAVGGGVTRR257 pKa = 11.84IQDD260 pKa = 3.39ATDD263 pKa = 3.59YY264 pKa = 10.69EE265 pKa = 5.05PNTTPPAGTIYY276 pKa = 10.61YY277 pKa = 9.82FSKK280 pKa = 10.95AIANFPEE287 pKa = 5.25DD288 pKa = 3.35WTCPDD293 pKa = 3.48RR294 pKa = 11.84GTYY297 pKa = 8.51STDD300 pKa = 3.31PGDD303 pKa = 3.61GTQYY307 pKa = 11.62NLARR311 pKa = 11.84YY312 pKa = 9.48DD313 pKa = 3.48RR314 pKa = 11.84DD315 pKa = 3.52GRR317 pKa = 11.84DD318 pKa = 3.46TQSVFPSSGSIWWTFDD334 pKa = 2.88LTQPWTQMTYY344 pKa = 9.99TGLGAQSICTANSRR358 pKa = 11.84KK359 pKa = 10.11AYY361 pKa = 10.28NGTVNGWPLDD371 pKa = 3.48GSTDD375 pKa = 3.56KK376 pKa = 10.85ILYY379 pKa = 9.13VALSDD384 pKa = 4.25LGTGIPIPLAAGDD397 pKa = 3.81YY398 pKa = 10.45LRR400 pKa = 11.84YY401 pKa = 10.32DD402 pKa = 3.71GGDD405 pKa = 3.5FRR407 pKa = 11.84PKK409 pKa = 10.18QLATVAEE416 pKa = 4.39TGAYY420 pKa = 9.74SDD422 pKa = 4.92LSGAPTLGTAAAAATTDD439 pKa = 3.71FATAAQGTLADD450 pKa = 4.26SAVQPTDD457 pKa = 4.55SIDD460 pKa = 3.35TLADD464 pKa = 3.39VTVTTPSNGEE474 pKa = 3.72ALVWNGSAWVNSTVSTVGSIDD495 pKa = 4.39DD496 pKa = 4.16LTDD499 pKa = 2.95VDD501 pKa = 4.5TTTSAPTNGQVLEE514 pKa = 4.37WNGTNWVPATIAAGGVTSIIAGTGISVDD542 pKa = 3.42QATGDD547 pKa = 3.57VTITATGGGVSGIGVATRR565 pKa = 11.84VSDD568 pKa = 3.48SGTAASGALALTGLGSSGMLHH589 pKa = 6.02TLSTDD594 pKa = 3.06LDD596 pKa = 3.65AWVVFYY602 pKa = 10.85GSAADD607 pKa = 3.93RR608 pKa = 11.84TADD611 pKa = 3.62AGRR614 pKa = 11.84AYY616 pKa = 9.92NTDD619 pKa = 3.53PAAGSGVLAEE629 pKa = 4.98AYY631 pKa = 7.75VTTAGAILFTPGTGYY646 pKa = 10.26MNNDD650 pKa = 3.26TTEE653 pKa = 3.82TAALYY658 pKa = 10.42LAVRR662 pKa = 11.84DD663 pKa = 3.85QAGAAVNATLTVTAYY678 pKa = 10.62VQGGYY683 pKa = 10.62DD684 pKa = 3.63GVSGGTFGSGG694 pKa = 2.79
MM1 pKa = 7.63AVPSNRR7 pKa = 11.84VPVRR11 pKa = 11.84VARR14 pKa = 11.84GLKK17 pKa = 10.04SALTANLADD26 pKa = 4.16LLEE29 pKa = 5.37GEE31 pKa = 4.59LVYY34 pKa = 11.13AKK36 pKa = 10.76DD37 pKa = 3.53EE38 pKa = 4.7DD39 pKa = 3.97KK40 pKa = 11.33LYY42 pKa = 10.04MIEE45 pKa = 4.46GGTLVAMGADD55 pKa = 3.96LAAASILDD63 pKa = 4.14LGDD66 pKa = 3.71VEE68 pKa = 5.32PNSLPDD74 pKa = 3.44GARR77 pKa = 11.84YY78 pKa = 9.82ALGGVQFPGNGLWYY92 pKa = 10.07RR93 pKa = 11.84SGSRR97 pKa = 11.84ISWGAIDD104 pKa = 5.55ADD106 pKa = 3.96STDD109 pKa = 3.42RR110 pKa = 11.84TSQITAASGSIFWSTDD126 pKa = 2.43GATWNEE132 pKa = 3.73IFFTGSPQTVGDD144 pKa = 4.21SYY146 pKa = 11.7YY147 pKa = 10.24IDD149 pKa = 3.44ITSGSLPALTGDD161 pKa = 4.62LYY163 pKa = 11.35FSFSTPGTPSPAPLADD179 pKa = 3.49GDD181 pKa = 4.05YY182 pKa = 11.26LRR184 pKa = 11.84FDD186 pKa = 3.68GTDD189 pKa = 3.52FRR191 pKa = 11.84PQQLATVAEE200 pKa = 4.39TGAYY204 pKa = 9.7SDD206 pKa = 5.03LSGTPTLVEE215 pKa = 4.8NIDD218 pKa = 4.18DD219 pKa = 4.12LTDD222 pKa = 3.25VDD224 pKa = 4.57TSTSAPTTDD233 pKa = 5.61QVLAWNGTNWVPSDD247 pKa = 3.42VSAVGGGVTRR257 pKa = 11.84IQDD260 pKa = 3.39ATDD263 pKa = 3.59YY264 pKa = 10.69EE265 pKa = 5.05PNTTPPAGTIYY276 pKa = 10.61YY277 pKa = 9.82FSKK280 pKa = 10.95AIANFPEE287 pKa = 5.25DD288 pKa = 3.35WTCPDD293 pKa = 3.48RR294 pKa = 11.84GTYY297 pKa = 8.51STDD300 pKa = 3.31PGDD303 pKa = 3.61GTQYY307 pKa = 11.62NLARR311 pKa = 11.84YY312 pKa = 9.48DD313 pKa = 3.48RR314 pKa = 11.84DD315 pKa = 3.52GRR317 pKa = 11.84DD318 pKa = 3.46TQSVFPSSGSIWWTFDD334 pKa = 2.88LTQPWTQMTYY344 pKa = 9.99TGLGAQSICTANSRR358 pKa = 11.84KK359 pKa = 10.11AYY361 pKa = 10.28NGTVNGWPLDD371 pKa = 3.48GSTDD375 pKa = 3.56KK376 pKa = 10.85ILYY379 pKa = 9.13VALSDD384 pKa = 4.25LGTGIPIPLAAGDD397 pKa = 3.81YY398 pKa = 10.45LRR400 pKa = 11.84YY401 pKa = 10.32DD402 pKa = 3.71GGDD405 pKa = 3.5FRR407 pKa = 11.84PKK409 pKa = 10.18QLATVAEE416 pKa = 4.39TGAYY420 pKa = 9.74SDD422 pKa = 4.92LSGAPTLGTAAAAATTDD439 pKa = 3.71FATAAQGTLADD450 pKa = 4.26SAVQPTDD457 pKa = 4.55SIDD460 pKa = 3.35TLADD464 pKa = 3.39VTVTTPSNGEE474 pKa = 3.72ALVWNGSAWVNSTVSTVGSIDD495 pKa = 4.39DD496 pKa = 4.16LTDD499 pKa = 2.95VDD501 pKa = 4.5TTTSAPTNGQVLEE514 pKa = 4.37WNGTNWVPATIAAGGVTSIIAGTGISVDD542 pKa = 3.42QATGDD547 pKa = 3.57VTITATGGGVSGIGVATRR565 pKa = 11.84VSDD568 pKa = 3.48SGTAASGALALTGLGSSGMLHH589 pKa = 6.02TLSTDD594 pKa = 3.06LDD596 pKa = 3.65AWVVFYY602 pKa = 10.85GSAADD607 pKa = 3.93RR608 pKa = 11.84TADD611 pKa = 3.62AGRR614 pKa = 11.84AYY616 pKa = 9.92NTDD619 pKa = 3.53PAAGSGVLAEE629 pKa = 4.98AYY631 pKa = 7.75VTTAGAILFTPGTGYY646 pKa = 10.26MNNDD650 pKa = 3.26TTEE653 pKa = 3.82TAALYY658 pKa = 10.42LAVRR662 pKa = 11.84DD663 pKa = 3.85QAGAAVNATLTVTAYY678 pKa = 10.62VQGGYY683 pKa = 10.62DD684 pKa = 3.63GVSGGTFGSGG694 pKa = 2.79
Molecular weight: 71.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4YCP0|F4YCP0_9CAUD Uncharacterized protein OS=Synechococcus phage S-CBS2 OX=753084 GN=S-CBS2_gp059 PE=4 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84LSHH5 pKa = 6.25QEE7 pKa = 3.49AEE9 pKa = 4.46ARR11 pKa = 11.84RR12 pKa = 11.84QLYY15 pKa = 10.18EE16 pKa = 4.05EE17 pKa = 4.54GGKK20 pKa = 10.51AIGEE24 pKa = 4.24LLCTYY29 pKa = 9.73PEE31 pKa = 4.68KK32 pKa = 10.75EE33 pKa = 3.94PQIWKK38 pKa = 9.68LVAGAGLWTRR48 pKa = 11.84KK49 pKa = 9.9DD50 pKa = 3.09LAAFKK55 pKa = 10.53SLWRR59 pKa = 11.84RR60 pKa = 11.84DD61 pKa = 3.48HH62 pKa = 5.97PVEE65 pKa = 3.79YY66 pKa = 10.75SRR68 pKa = 11.84IKK70 pKa = 8.61QARR73 pKa = 11.84YY74 pKa = 9.66DD75 pKa = 3.81RR76 pKa = 11.84KK77 pKa = 9.29QLARR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84AAKK86 pKa = 10.32LEE88 pKa = 3.94KK89 pKa = 9.71TRR91 pKa = 11.84EE92 pKa = 3.97ASKK95 pKa = 9.58PQAMPTEE102 pKa = 4.05RR103 pKa = 11.84RR104 pKa = 11.84PRR106 pKa = 11.84KK107 pKa = 9.61DD108 pKa = 3.1LRR110 pKa = 11.84DD111 pKa = 3.61RR112 pKa = 11.84KK113 pKa = 10.46
MM1 pKa = 7.64RR2 pKa = 11.84LSHH5 pKa = 6.25QEE7 pKa = 3.49AEE9 pKa = 4.46ARR11 pKa = 11.84RR12 pKa = 11.84QLYY15 pKa = 10.18EE16 pKa = 4.05EE17 pKa = 4.54GGKK20 pKa = 10.51AIGEE24 pKa = 4.24LLCTYY29 pKa = 9.73PEE31 pKa = 4.68KK32 pKa = 10.75EE33 pKa = 3.94PQIWKK38 pKa = 9.68LVAGAGLWTRR48 pKa = 11.84KK49 pKa = 9.9DD50 pKa = 3.09LAAFKK55 pKa = 10.53SLWRR59 pKa = 11.84RR60 pKa = 11.84DD61 pKa = 3.48HH62 pKa = 5.97PVEE65 pKa = 3.79YY66 pKa = 10.75SRR68 pKa = 11.84IKK70 pKa = 8.61QARR73 pKa = 11.84YY74 pKa = 9.66DD75 pKa = 3.81RR76 pKa = 11.84KK77 pKa = 9.29QLARR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84AAKK86 pKa = 10.32LEE88 pKa = 3.94KK89 pKa = 9.71TRR91 pKa = 11.84EE92 pKa = 3.97ASKK95 pKa = 9.58PQAMPTEE102 pKa = 4.05RR103 pKa = 11.84RR104 pKa = 11.84PRR106 pKa = 11.84KK107 pKa = 9.61DD108 pKa = 3.1LRR110 pKa = 11.84DD111 pKa = 3.61RR112 pKa = 11.84KK113 pKa = 10.46
Molecular weight: 13.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
21656 |
43 |
956 |
212.3 |
23.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.656 ± 0.327 | 1.261 ± 0.274 |
6.008 ± 0.17 | 6.248 ± 0.321 |
3.325 ± 0.146 | 9.138 ± 0.525 |
1.598 ± 0.169 | 5.042 ± 0.18 |
4.738 ± 0.334 | 8.016 ± 0.262 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.912 ± 0.14 | 4.248 ± 0.198 |
4.308 ± 0.203 | 4.059 ± 0.246 |
5.052 ± 0.311 | 6.968 ± 0.335 |
6.686 ± 0.414 | 6.516 ± 0.226 |
1.635 ± 0.106 | 3.588 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
