
Citrus vein enation virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Enamovirus
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
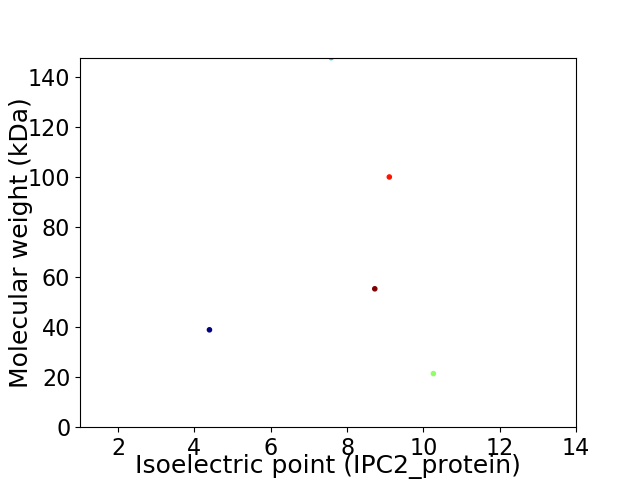
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S0EQ55|S0EQ55_9LUTE Serine protease OS=Citrus vein enation virus OX=1301220 PE=4 SV=1
MM1 pKa = 7.64PCYY4 pKa = 10.06HH5 pKa = 7.28VIPYY9 pKa = 10.42DD10 pKa = 3.63YY11 pKa = 9.78FQPIHH16 pKa = 6.42SSNWLGASLLALINYY31 pKa = 9.33VGMGCPTQEE40 pKa = 3.97GTPCEE45 pKa = 4.32EE46 pKa = 4.44PTCVALPYY54 pKa = 10.84VLLCALALLWGHH66 pKa = 6.48PRR68 pKa = 11.84SCGRR72 pKa = 11.84RR73 pKa = 11.84GGVSLEE79 pKa = 3.86ALTLPRR85 pKa = 11.84GFNIVGPPDD94 pKa = 3.6FLWYY98 pKa = 10.28FRR100 pKa = 11.84ICAARR105 pKa = 11.84IGLIIPQSYY114 pKa = 8.36WRR116 pKa = 11.84DD117 pKa = 3.49DD118 pKa = 3.31GRR120 pKa = 11.84RR121 pKa = 11.84AIPLTHH127 pKa = 6.72CALFTSLPCWLMAASGKK144 pKa = 10.4AEE146 pKa = 4.05YY147 pKa = 10.02HH148 pKa = 5.56CQGYY152 pKa = 8.25VAPEE156 pKa = 3.83LQLPGDD162 pKa = 4.34GPEE165 pKa = 3.82HH166 pKa = 5.32TVWVEE171 pKa = 3.85EE172 pKa = 5.09KK173 pKa = 8.74WACWHH178 pKa = 5.74EE179 pKa = 4.59LIVGLLGTHH188 pKa = 6.11TNVASILRR196 pKa = 11.84VLPDD200 pKa = 3.5GGVTPHH206 pKa = 6.04SQVLGKK212 pKa = 10.62RR213 pKa = 11.84NLIVDD218 pKa = 4.55AFTSISTMAVGTDD231 pKa = 3.31QPPPNRR237 pKa = 11.84YY238 pKa = 8.58MDD240 pKa = 4.31DD241 pKa = 3.74CSLTRR246 pKa = 11.84LFCLRR251 pKa = 11.84EE252 pKa = 4.17PYY254 pKa = 10.7LSGYY258 pKa = 8.88DD259 pKa = 3.37TDD261 pKa = 4.28NFSPGLHH268 pKa = 5.96ITLPWPEE275 pKa = 3.84IPEE278 pKa = 4.3GGEE281 pKa = 3.85YY282 pKa = 10.11LAHH285 pKa = 6.77SFAEE289 pKa = 4.14LDD291 pKa = 3.69QEE293 pKa = 4.62FADD296 pKa = 4.69LLAHH300 pKa = 6.38LHH302 pKa = 5.15VHH304 pKa = 7.21DD305 pKa = 5.74DD306 pKa = 4.04EE307 pKa = 6.17EE308 pKa = 5.35DD309 pKa = 3.72LGWFSDD315 pKa = 3.98DD316 pKa = 4.29GEE318 pKa = 5.01GDD320 pKa = 3.42IEE322 pKa = 4.92NPPAPHH328 pKa = 6.1NTDD331 pKa = 3.38EE332 pKa = 4.52EE333 pKa = 4.77DD334 pKa = 4.93FGDD337 pKa = 4.94SDD339 pKa = 5.54DD340 pKa = 5.18DD341 pKa = 5.28DD342 pKa = 6.75SIGGNAGSGPEE353 pKa = 3.76AA354 pKa = 4.5
MM1 pKa = 7.64PCYY4 pKa = 10.06HH5 pKa = 7.28VIPYY9 pKa = 10.42DD10 pKa = 3.63YY11 pKa = 9.78FQPIHH16 pKa = 6.42SSNWLGASLLALINYY31 pKa = 9.33VGMGCPTQEE40 pKa = 3.97GTPCEE45 pKa = 4.32EE46 pKa = 4.44PTCVALPYY54 pKa = 10.84VLLCALALLWGHH66 pKa = 6.48PRR68 pKa = 11.84SCGRR72 pKa = 11.84RR73 pKa = 11.84GGVSLEE79 pKa = 3.86ALTLPRR85 pKa = 11.84GFNIVGPPDD94 pKa = 3.6FLWYY98 pKa = 10.28FRR100 pKa = 11.84ICAARR105 pKa = 11.84IGLIIPQSYY114 pKa = 8.36WRR116 pKa = 11.84DD117 pKa = 3.49DD118 pKa = 3.31GRR120 pKa = 11.84RR121 pKa = 11.84AIPLTHH127 pKa = 6.72CALFTSLPCWLMAASGKK144 pKa = 10.4AEE146 pKa = 4.05YY147 pKa = 10.02HH148 pKa = 5.56CQGYY152 pKa = 8.25VAPEE156 pKa = 3.83LQLPGDD162 pKa = 4.34GPEE165 pKa = 3.82HH166 pKa = 5.32TVWVEE171 pKa = 3.85EE172 pKa = 5.09KK173 pKa = 8.74WACWHH178 pKa = 5.74EE179 pKa = 4.59LIVGLLGTHH188 pKa = 6.11TNVASILRR196 pKa = 11.84VLPDD200 pKa = 3.5GGVTPHH206 pKa = 6.04SQVLGKK212 pKa = 10.62RR213 pKa = 11.84NLIVDD218 pKa = 4.55AFTSISTMAVGTDD231 pKa = 3.31QPPPNRR237 pKa = 11.84YY238 pKa = 8.58MDD240 pKa = 4.31DD241 pKa = 3.74CSLTRR246 pKa = 11.84LFCLRR251 pKa = 11.84EE252 pKa = 4.17PYY254 pKa = 10.7LSGYY258 pKa = 8.88DD259 pKa = 3.37TDD261 pKa = 4.28NFSPGLHH268 pKa = 5.96ITLPWPEE275 pKa = 3.84IPEE278 pKa = 4.3GGEE281 pKa = 3.85YY282 pKa = 10.11LAHH285 pKa = 6.77SFAEE289 pKa = 4.14LDD291 pKa = 3.69QEE293 pKa = 4.62FADD296 pKa = 4.69LLAHH300 pKa = 6.38LHH302 pKa = 5.15VHH304 pKa = 7.21DD305 pKa = 5.74DD306 pKa = 4.04EE307 pKa = 6.17EE308 pKa = 5.35DD309 pKa = 3.72LGWFSDD315 pKa = 3.98DD316 pKa = 4.29GEE318 pKa = 5.01GDD320 pKa = 3.42IEE322 pKa = 4.92NPPAPHH328 pKa = 6.1NTDD331 pKa = 3.38EE332 pKa = 4.52EE333 pKa = 4.77DD334 pKa = 4.93FGDD337 pKa = 4.94SDD339 pKa = 5.54DD340 pKa = 5.18DD341 pKa = 5.28DD342 pKa = 6.75SIGGNAGSGPEE353 pKa = 3.76AA354 pKa = 4.5
Molecular weight: 38.96 kDa
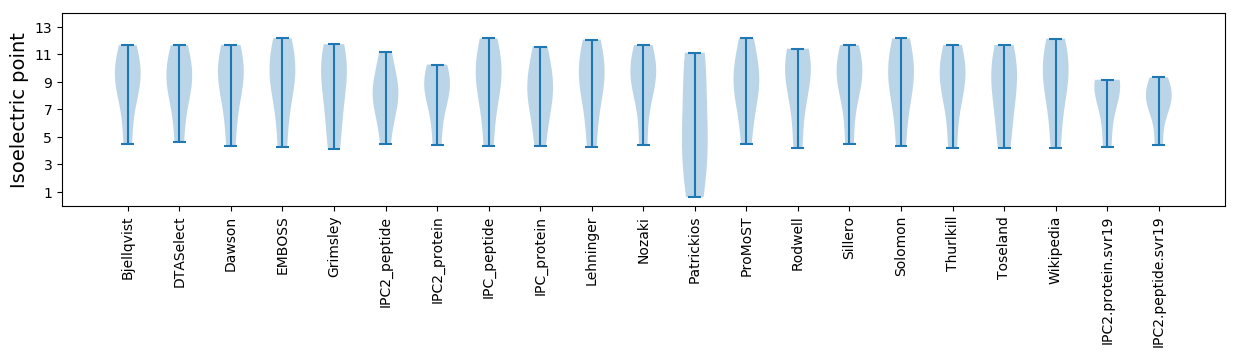
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S0ERJ8|S0ERJ8_9LUTE CP OS=Citrus vein enation virus OX=1301220 PE=4 SV=1
MM1 pKa = 7.36VSRR4 pKa = 11.84NQRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84NRR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84MRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84QAPNRR23 pKa = 11.84VVVMGNNPTRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84NAPARR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84AVNRR49 pKa = 11.84STGEE53 pKa = 3.5MRR55 pKa = 11.84PYY57 pKa = 10.38HH58 pKa = 7.16LYY60 pKa = 10.83GLKK63 pKa = 10.92CNDD66 pKa = 3.19KK67 pKa = 11.03GYY69 pKa = 8.96LTFGPSGQTPSLGGGILKK87 pKa = 10.48AFSEE91 pKa = 4.48YY92 pKa = 10.58KK93 pKa = 8.8ITQLRR98 pKa = 11.84VQWKK102 pKa = 8.54AQASSTAAGSMAIQIGLGTSLTALDD127 pKa = 3.68NRR129 pKa = 11.84AISFKK134 pKa = 9.98LTSSGQRR141 pKa = 11.84VFTARR146 pKa = 11.84DD147 pKa = 3.21LGGDD151 pKa = 2.87GRR153 pKa = 11.84MYY155 pKa = 10.93NSSNEE160 pKa = 3.99DD161 pKa = 3.32QFRR164 pKa = 11.84LAYY167 pKa = 9.74QGNGDD172 pKa = 3.82STLGGDD178 pKa = 4.17LLCFFRR184 pKa = 11.84LEE186 pKa = 4.05TRR188 pKa = 11.84LPKK191 pKa = 10.53
MM1 pKa = 7.36VSRR4 pKa = 11.84NQRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84NRR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84MRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84QAPNRR23 pKa = 11.84VVVMGNNPTRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84NAPARR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84AVNRR49 pKa = 11.84STGEE53 pKa = 3.5MRR55 pKa = 11.84PYY57 pKa = 10.38HH58 pKa = 7.16LYY60 pKa = 10.83GLKK63 pKa = 10.92CNDD66 pKa = 3.19KK67 pKa = 11.03GYY69 pKa = 8.96LTFGPSGQTPSLGGGILKK87 pKa = 10.48AFSEE91 pKa = 4.48YY92 pKa = 10.58KK93 pKa = 8.8ITQLRR98 pKa = 11.84VQWKK102 pKa = 8.54AQASSTAAGSMAIQIGLGTSLTALDD127 pKa = 3.68NRR129 pKa = 11.84AISFKK134 pKa = 9.98LTSSGQRR141 pKa = 11.84VFTARR146 pKa = 11.84DD147 pKa = 3.21LGGDD151 pKa = 2.87GRR153 pKa = 11.84MYY155 pKa = 10.93NSSNEE160 pKa = 3.99DD161 pKa = 3.32QFRR164 pKa = 11.84LAYY167 pKa = 9.74QGNGDD172 pKa = 3.82STLGGDD178 pKa = 4.17LLCFFRR184 pKa = 11.84LEE186 pKa = 4.05TRR188 pKa = 11.84LPKK191 pKa = 10.53
Molecular weight: 21.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3264 |
191 |
1323 |
652.8 |
72.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.261 ± 0.106 | 2.145 ± 0.307 |
4.504 ± 0.537 | 5.637 ± 0.384 |
3.37 ± 0.224 | 7.629 ± 0.443 |
1.777 ± 0.39 | 4.289 ± 0.199 |
4.626 ± 0.594 | 10.968 ± 1.007 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.604 ± 0.347 | 3.401 ± 0.385 |
5.974 ± 0.584 | 3.676 ± 0.238 |
7.414 ± 1.002 | 8.027 ± 0.52 |
5.392 ± 0.208 | 6.342 ± 0.509 |
2.022 ± 0.237 | 2.911 ± 0.247 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
