
Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanospirillaceae; Methanospirillum; Methanospirillum hungatei
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
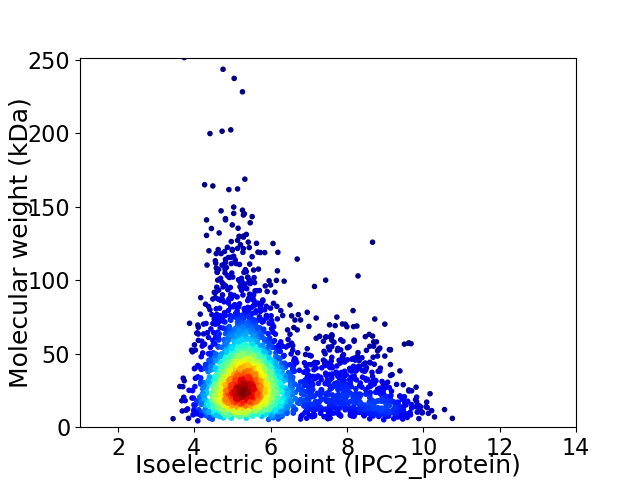
Virtual 2D-PAGE plot for 3087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q2FS66|SYR_METHJ Arginine--tRNA ligase OS=Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1) OX=323259 GN=argS PE=3 SV=1
MM1 pKa = 7.43RR2 pKa = 11.84AAHH5 pKa = 6.56IFVLLLLLWITPAGMGEE22 pKa = 4.08EE23 pKa = 4.33TQDD26 pKa = 4.92DD27 pKa = 3.77ISPAQTHH34 pKa = 6.08NLLVIYY40 pKa = 8.28MIAGDD45 pKa = 5.22LEE47 pKa = 4.69TEE49 pKa = 4.38TQSATMNIAEE59 pKa = 4.64ILDD62 pKa = 4.55GYY64 pKa = 11.14GKK66 pKa = 9.05TSEE69 pKa = 4.13QNLQIVLGYY78 pKa = 10.23GGSKK82 pKa = 9.15TPGFGGITYY91 pKa = 7.72VTVKK95 pKa = 10.65EE96 pKa = 4.07LMEE99 pKa = 4.81DD100 pKa = 3.36ASDD103 pKa = 3.85GVIGNTEE110 pKa = 3.93DD111 pKa = 4.14VLYY114 pKa = 10.32HH115 pKa = 7.46DD116 pKa = 5.86PDD118 pKa = 5.31ADD120 pKa = 3.68MGDD123 pKa = 3.83KK124 pKa = 9.67KK125 pKa = 9.47TLEE128 pKa = 4.85HH129 pKa = 5.87FLSWIGEE136 pKa = 4.17NFPSDD141 pKa = 3.33RR142 pKa = 11.84KK143 pKa = 10.42ILVFWDD149 pKa = 3.13HH150 pKa = 6.48GGGYY154 pKa = 10.35DD155 pKa = 3.74GFGVDD160 pKa = 4.86EE161 pKa = 4.42VTDD164 pKa = 4.3GQLSLTDD171 pKa = 3.43ISQALEE177 pKa = 4.19ASGSTYY183 pKa = 11.4DD184 pKa = 4.72LIGYY188 pKa = 7.57DD189 pKa = 3.76ACLMGALEE197 pKa = 4.29VAKK200 pKa = 10.79AMEE203 pKa = 4.84PYY205 pKa = 10.38GILLIGSEE213 pKa = 4.27EE214 pKa = 4.34TEE216 pKa = 4.46PGTGWEE222 pKa = 4.13YY223 pKa = 9.81EE224 pKa = 4.15TWIKK228 pKa = 11.03ALGEE232 pKa = 3.91NPEE235 pKa = 4.08IGFEE239 pKa = 4.01EE240 pKa = 4.61LGQIIVDD247 pKa = 3.94TYY249 pKa = 10.51MNRR252 pKa = 11.84DD253 pKa = 3.58DD254 pKa = 3.73TGKK257 pKa = 7.73TLSVIDD263 pKa = 3.91LTKK266 pKa = 10.38IPALIQALDD275 pKa = 3.61RR276 pKa = 11.84LGTSMMPYY284 pKa = 8.58TEE286 pKa = 5.01SIEE289 pKa = 4.33GYY291 pKa = 9.35RR292 pKa = 11.84VVGKK296 pKa = 10.35AYY298 pKa = 9.66QVPARR303 pKa = 11.84YY304 pKa = 9.77GSDD307 pKa = 2.85NRR309 pKa = 11.84EE310 pKa = 4.02GGEE313 pKa = 3.97TSVDD317 pKa = 3.22LKK319 pKa = 11.45SFLKK323 pKa = 10.91AIGNQTPDD331 pKa = 3.59LADD334 pKa = 4.68EE335 pKa = 4.39IAVVTTRR342 pKa = 11.84IDD344 pKa = 3.41EE345 pKa = 4.34TVLYY349 pKa = 9.81HH350 pKa = 7.5RR351 pKa = 11.84NDD353 pKa = 3.25EE354 pKa = 4.3YY355 pKa = 11.66VPEE358 pKa = 4.64SGGLSIMSPSRR369 pKa = 11.84ITPDD373 pKa = 3.0LYY375 pKa = 11.35EE376 pKa = 4.44EE377 pKa = 4.98LGDD380 pKa = 4.47DD381 pKa = 5.05ARR383 pKa = 11.84ITPGWDD389 pKa = 2.98SFFVNLLEE397 pKa = 5.49ISGQDD402 pKa = 3.4TEE404 pKa = 4.75KK405 pKa = 10.92PEE407 pKa = 3.99IMRR410 pKa = 11.84SDD412 pKa = 3.36SGFLIEE418 pKa = 5.99DD419 pKa = 3.76PTNTASVYY427 pKa = 10.79AEE429 pKa = 4.17YY430 pKa = 10.9YY431 pKa = 9.52YY432 pKa = 11.49ADD434 pKa = 3.95GDD436 pKa = 4.06EE437 pKa = 5.81LILLGNEE444 pKa = 4.13PLDD447 pKa = 3.98PDD449 pKa = 3.77EE450 pKa = 4.84NGEE453 pKa = 4.2YY454 pKa = 9.94TLPEE458 pKa = 3.75WDD460 pKa = 3.59GRR462 pKa = 11.84WYY464 pKa = 10.86YY465 pKa = 10.17LQDD468 pKa = 3.47MNNEE472 pKa = 4.0DD473 pKa = 3.93NYY475 pKa = 11.86ALLGMNFEE483 pKa = 5.19SIGPSGSVLYY493 pKa = 10.16TSEE496 pKa = 3.77IDD498 pKa = 5.08LIRR501 pKa = 11.84EE502 pKa = 4.28SLNTTAVLNVYY513 pKa = 10.23INPQTGEE520 pKa = 4.1TRR522 pKa = 11.84LVACPYY528 pKa = 9.32TIRR531 pKa = 11.84PNGIIQFSRR540 pKa = 11.84QNLDD544 pKa = 3.64LEE546 pKa = 4.98PNDD549 pKa = 3.89TVYY552 pKa = 11.04SYY554 pKa = 11.22AWKK557 pKa = 10.34VDD559 pKa = 3.0EE560 pKa = 4.45DD561 pKa = 4.68TEE563 pKa = 4.68TGGEE567 pKa = 4.02WVEE570 pKa = 4.17IGALVISGNTKK581 pKa = 10.51LIYY584 pKa = 10.34DD585 pKa = 4.41ILPDD589 pKa = 3.51GTYY592 pKa = 10.82AQALYY597 pKa = 10.86AEE599 pKa = 4.95YY600 pKa = 10.92GNKK603 pKa = 9.81PGDD606 pKa = 3.63YY607 pKa = 10.93AGMQVFQIEE616 pKa = 4.34NGEE619 pKa = 3.97ITLVEE624 pKa = 4.31SEE626 pKa = 4.58TNITALQEE634 pKa = 4.15PEE636 pKa = 4.51EE637 pKa = 4.28NNQQ640 pKa = 3.05
MM1 pKa = 7.43RR2 pKa = 11.84AAHH5 pKa = 6.56IFVLLLLLWITPAGMGEE22 pKa = 4.08EE23 pKa = 4.33TQDD26 pKa = 4.92DD27 pKa = 3.77ISPAQTHH34 pKa = 6.08NLLVIYY40 pKa = 8.28MIAGDD45 pKa = 5.22LEE47 pKa = 4.69TEE49 pKa = 4.38TQSATMNIAEE59 pKa = 4.64ILDD62 pKa = 4.55GYY64 pKa = 11.14GKK66 pKa = 9.05TSEE69 pKa = 4.13QNLQIVLGYY78 pKa = 10.23GGSKK82 pKa = 9.15TPGFGGITYY91 pKa = 7.72VTVKK95 pKa = 10.65EE96 pKa = 4.07LMEE99 pKa = 4.81DD100 pKa = 3.36ASDD103 pKa = 3.85GVIGNTEE110 pKa = 3.93DD111 pKa = 4.14VLYY114 pKa = 10.32HH115 pKa = 7.46DD116 pKa = 5.86PDD118 pKa = 5.31ADD120 pKa = 3.68MGDD123 pKa = 3.83KK124 pKa = 9.67KK125 pKa = 9.47TLEE128 pKa = 4.85HH129 pKa = 5.87FLSWIGEE136 pKa = 4.17NFPSDD141 pKa = 3.33RR142 pKa = 11.84KK143 pKa = 10.42ILVFWDD149 pKa = 3.13HH150 pKa = 6.48GGGYY154 pKa = 10.35DD155 pKa = 3.74GFGVDD160 pKa = 4.86EE161 pKa = 4.42VTDD164 pKa = 4.3GQLSLTDD171 pKa = 3.43ISQALEE177 pKa = 4.19ASGSTYY183 pKa = 11.4DD184 pKa = 4.72LIGYY188 pKa = 7.57DD189 pKa = 3.76ACLMGALEE197 pKa = 4.29VAKK200 pKa = 10.79AMEE203 pKa = 4.84PYY205 pKa = 10.38GILLIGSEE213 pKa = 4.27EE214 pKa = 4.34TEE216 pKa = 4.46PGTGWEE222 pKa = 4.13YY223 pKa = 9.81EE224 pKa = 4.15TWIKK228 pKa = 11.03ALGEE232 pKa = 3.91NPEE235 pKa = 4.08IGFEE239 pKa = 4.01EE240 pKa = 4.61LGQIIVDD247 pKa = 3.94TYY249 pKa = 10.51MNRR252 pKa = 11.84DD253 pKa = 3.58DD254 pKa = 3.73TGKK257 pKa = 7.73TLSVIDD263 pKa = 3.91LTKK266 pKa = 10.38IPALIQALDD275 pKa = 3.61RR276 pKa = 11.84LGTSMMPYY284 pKa = 8.58TEE286 pKa = 5.01SIEE289 pKa = 4.33GYY291 pKa = 9.35RR292 pKa = 11.84VVGKK296 pKa = 10.35AYY298 pKa = 9.66QVPARR303 pKa = 11.84YY304 pKa = 9.77GSDD307 pKa = 2.85NRR309 pKa = 11.84EE310 pKa = 4.02GGEE313 pKa = 3.97TSVDD317 pKa = 3.22LKK319 pKa = 11.45SFLKK323 pKa = 10.91AIGNQTPDD331 pKa = 3.59LADD334 pKa = 4.68EE335 pKa = 4.39IAVVTTRR342 pKa = 11.84IDD344 pKa = 3.41EE345 pKa = 4.34TVLYY349 pKa = 9.81HH350 pKa = 7.5RR351 pKa = 11.84NDD353 pKa = 3.25EE354 pKa = 4.3YY355 pKa = 11.66VPEE358 pKa = 4.64SGGLSIMSPSRR369 pKa = 11.84ITPDD373 pKa = 3.0LYY375 pKa = 11.35EE376 pKa = 4.44EE377 pKa = 4.98LGDD380 pKa = 4.47DD381 pKa = 5.05ARR383 pKa = 11.84ITPGWDD389 pKa = 2.98SFFVNLLEE397 pKa = 5.49ISGQDD402 pKa = 3.4TEE404 pKa = 4.75KK405 pKa = 10.92PEE407 pKa = 3.99IMRR410 pKa = 11.84SDD412 pKa = 3.36SGFLIEE418 pKa = 5.99DD419 pKa = 3.76PTNTASVYY427 pKa = 10.79AEE429 pKa = 4.17YY430 pKa = 10.9YY431 pKa = 9.52YY432 pKa = 11.49ADD434 pKa = 3.95GDD436 pKa = 4.06EE437 pKa = 5.81LILLGNEE444 pKa = 4.13PLDD447 pKa = 3.98PDD449 pKa = 3.77EE450 pKa = 4.84NGEE453 pKa = 4.2YY454 pKa = 9.94TLPEE458 pKa = 3.75WDD460 pKa = 3.59GRR462 pKa = 11.84WYY464 pKa = 10.86YY465 pKa = 10.17LQDD468 pKa = 3.47MNNEE472 pKa = 4.0DD473 pKa = 3.93NYY475 pKa = 11.86ALLGMNFEE483 pKa = 5.19SIGPSGSVLYY493 pKa = 10.16TSEE496 pKa = 3.77IDD498 pKa = 5.08LIRR501 pKa = 11.84EE502 pKa = 4.28SLNTTAVLNVYY513 pKa = 10.23INPQTGEE520 pKa = 4.1TRR522 pKa = 11.84LVACPYY528 pKa = 9.32TIRR531 pKa = 11.84PNGIIQFSRR540 pKa = 11.84QNLDD544 pKa = 3.64LEE546 pKa = 4.98PNDD549 pKa = 3.89TVYY552 pKa = 11.04SYY554 pKa = 11.22AWKK557 pKa = 10.34VDD559 pKa = 3.0EE560 pKa = 4.45DD561 pKa = 4.68TEE563 pKa = 4.68TGGEE567 pKa = 4.02WVEE570 pKa = 4.17IGALVISGNTKK581 pKa = 10.51LIYY584 pKa = 10.34DD585 pKa = 4.41ILPDD589 pKa = 3.51GTYY592 pKa = 10.82AQALYY597 pKa = 10.86AEE599 pKa = 4.95YY600 pKa = 10.92GNKK603 pKa = 9.81PGDD606 pKa = 3.63YY607 pKa = 10.93AGMQVFQIEE616 pKa = 4.34NGEE619 pKa = 3.97ITLVEE624 pKa = 4.31SEE626 pKa = 4.58TNITALQEE634 pKa = 4.15PEE636 pKa = 4.51EE637 pKa = 4.28NNQQ640 pKa = 3.05
Molecular weight: 70.73 kDa
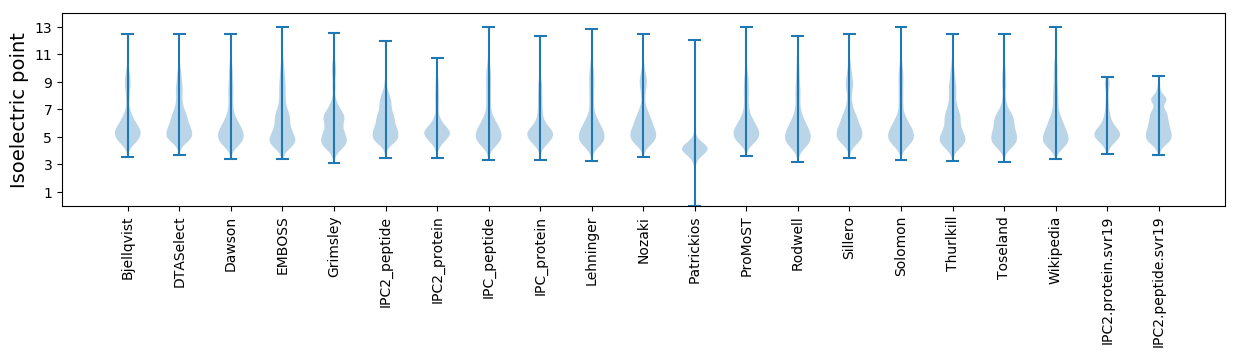
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2FTM5|Q2FTM5_METHJ Oligosaccharyl transferase OS=Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1) OX=323259 GN=Mhun_2859 PE=3 SV=1
MM1 pKa = 7.72SKK3 pKa = 8.9LTKK6 pKa = 9.67CRR8 pKa = 11.84KK9 pKa = 8.99IRR11 pKa = 11.84LSKK14 pKa = 9.95KK15 pKa = 5.62TTQNRR20 pKa = 11.84RR21 pKa = 11.84VPQWVMVKK29 pKa = 7.88TARR32 pKa = 11.84AVMAHH37 pKa = 6.04PQRR40 pKa = 11.84HH41 pKa = 4.16SWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.66VV50 pKa = 3.07
MM1 pKa = 7.72SKK3 pKa = 8.9LTKK6 pKa = 9.67CRR8 pKa = 11.84KK9 pKa = 8.99IRR11 pKa = 11.84LSKK14 pKa = 9.95KK15 pKa = 5.62TTQNRR20 pKa = 11.84RR21 pKa = 11.84VPQWVMVKK29 pKa = 7.88TARR32 pKa = 11.84AVMAHH37 pKa = 6.04PQRR40 pKa = 11.84HH41 pKa = 4.16SWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.66VV50 pKa = 3.07
Molecular weight: 6.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996237 |
41 |
2353 |
322.7 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.034 ± 0.047 | 1.366 ± 0.022 |
5.781 ± 0.033 | 6.755 ± 0.056 |
3.981 ± 0.034 | 7.352 ± 0.042 |
2.202 ± 0.021 | 8.521 ± 0.051 |
4.99 ± 0.041 | 9.18 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.637 ± 0.02 | 3.579 ± 0.033 |
4.686 ± 0.041 | 3.185 ± 0.025 |
5.179 ± 0.039 | 6.687 ± 0.043 |
5.803 ± 0.049 | 6.584 ± 0.037 |
1.06 ± 0.017 | 3.438 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
