
Po-Circo-like virus 41
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
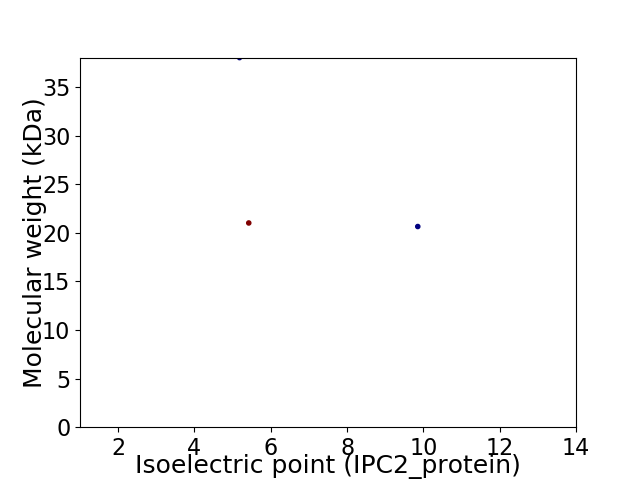
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
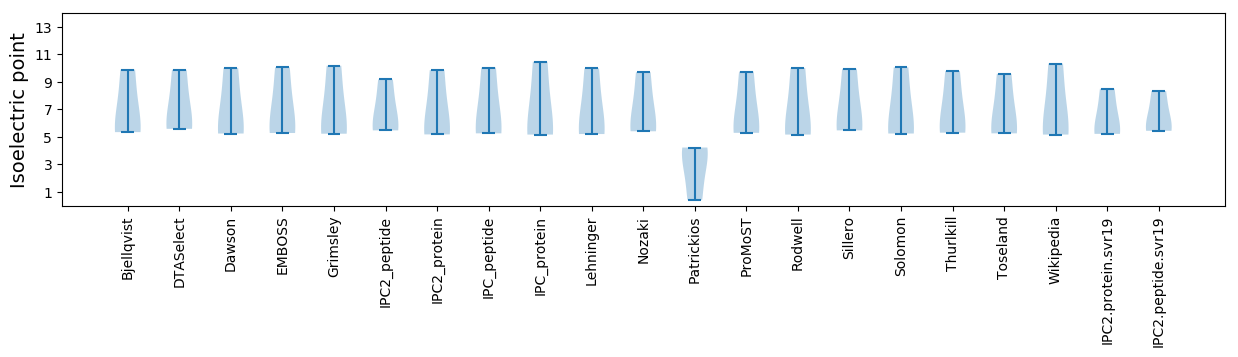
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8E3Y2|G8E3Y2_9CIRC Uncharacterized protein OS=Po-Circo-like virus 41 OX=1105385 PE=4 SV=1
MM1 pKa = 7.21SRR3 pKa = 11.84IRR5 pKa = 11.84GTRR8 pKa = 11.84WCFTINNFTQEE19 pKa = 3.97EE20 pKa = 4.59EE21 pKa = 4.23EE22 pKa = 4.67LVMNMPSTGWVKK34 pKa = 11.3SMIAEE39 pKa = 4.11EE40 pKa = 3.94EE41 pKa = 4.27HH42 pKa = 7.39LFEE45 pKa = 4.66GTPHH49 pKa = 5.35IQGYY53 pKa = 7.13FTTTKK58 pKa = 9.76RR59 pKa = 11.84TDD61 pKa = 3.55FTALTKK67 pKa = 10.19FFNGRR72 pKa = 11.84AHH74 pKa = 7.45LEE76 pKa = 3.88QAKK79 pKa = 10.58GSTRR83 pKa = 11.84DD84 pKa = 2.9NWNYY88 pKa = 9.98CSKK91 pKa = 9.81EE92 pKa = 3.74GNVIVEE98 pKa = 4.73FNRR101 pKa = 11.84PSTVDD106 pKa = 3.48RR107 pKa = 11.84VPHH110 pKa = 6.63RR111 pKa = 11.84AHH113 pKa = 7.43EE114 pKa = 4.36DD115 pKa = 3.71PKK117 pKa = 11.51DD118 pKa = 3.34EE119 pKa = 4.39WADD122 pKa = 4.76LMIQDD127 pKa = 4.9CEE129 pKa = 4.29QMDD132 pKa = 3.84RR133 pKa = 11.84EE134 pKa = 4.71TFRR137 pKa = 11.84SEE139 pKa = 3.96HH140 pKa = 6.63PNFFLNQRR148 pKa = 11.84DD149 pKa = 3.75KK150 pKa = 11.6YY151 pKa = 10.36EE152 pKa = 4.72RR153 pKa = 11.84IHH155 pKa = 7.16NEE157 pKa = 3.34YY158 pKa = 10.2LARR161 pKa = 11.84TTTVFDD167 pKa = 4.51GRR169 pKa = 11.84LHH171 pKa = 7.42DD172 pKa = 4.25KK173 pKa = 10.79NIWIYY178 pKa = 10.8GPPGSGKK185 pKa = 8.61STLARR190 pKa = 11.84SDD192 pKa = 3.45TPIDD196 pKa = 3.87HH197 pKa = 7.38IYY199 pKa = 10.48FKK201 pKa = 10.78PVDD204 pKa = 3.66KK205 pKa = 10.17WWDD208 pKa = 3.37GFDD211 pKa = 3.55PSKK214 pKa = 10.86HH215 pKa = 5.28NRR217 pKa = 11.84IVMDD221 pKa = 4.89DD222 pKa = 4.06YY223 pKa = 11.57PNEE226 pKa = 4.12ATGGAIFTQRR236 pKa = 11.84MKK238 pKa = 10.35IWGDD242 pKa = 2.94RR243 pKa = 11.84TTEE246 pKa = 3.76PRR248 pKa = 11.84PVKK251 pKa = 10.53GGTITVDD258 pKa = 3.2PNIPFIVTSNFPIDD272 pKa = 3.42EE273 pKa = 4.47CFSNDD278 pKa = 2.85VDD280 pKa = 3.75RR281 pKa = 11.84EE282 pKa = 4.28AIHH285 pKa = 7.16RR286 pKa = 11.84RR287 pKa = 11.84FQEE290 pKa = 3.78IYY292 pKa = 10.97LDD294 pKa = 3.97GDD296 pKa = 3.32KK297 pKa = 11.37SKK299 pKa = 11.27LDD301 pKa = 3.21RR302 pKa = 11.84FMHH305 pKa = 6.42LKK307 pKa = 10.61NLINPEE313 pKa = 4.48DD314 pKa = 4.63PLDD317 pKa = 4.58CDD319 pKa = 4.6DD320 pKa = 6.25SIVSDD325 pKa = 3.69
MM1 pKa = 7.21SRR3 pKa = 11.84IRR5 pKa = 11.84GTRR8 pKa = 11.84WCFTINNFTQEE19 pKa = 3.97EE20 pKa = 4.59EE21 pKa = 4.23EE22 pKa = 4.67LVMNMPSTGWVKK34 pKa = 11.3SMIAEE39 pKa = 4.11EE40 pKa = 3.94EE41 pKa = 4.27HH42 pKa = 7.39LFEE45 pKa = 4.66GTPHH49 pKa = 5.35IQGYY53 pKa = 7.13FTTTKK58 pKa = 9.76RR59 pKa = 11.84TDD61 pKa = 3.55FTALTKK67 pKa = 10.19FFNGRR72 pKa = 11.84AHH74 pKa = 7.45LEE76 pKa = 3.88QAKK79 pKa = 10.58GSTRR83 pKa = 11.84DD84 pKa = 2.9NWNYY88 pKa = 9.98CSKK91 pKa = 9.81EE92 pKa = 3.74GNVIVEE98 pKa = 4.73FNRR101 pKa = 11.84PSTVDD106 pKa = 3.48RR107 pKa = 11.84VPHH110 pKa = 6.63RR111 pKa = 11.84AHH113 pKa = 7.43EE114 pKa = 4.36DD115 pKa = 3.71PKK117 pKa = 11.51DD118 pKa = 3.34EE119 pKa = 4.39WADD122 pKa = 4.76LMIQDD127 pKa = 4.9CEE129 pKa = 4.29QMDD132 pKa = 3.84RR133 pKa = 11.84EE134 pKa = 4.71TFRR137 pKa = 11.84SEE139 pKa = 3.96HH140 pKa = 6.63PNFFLNQRR148 pKa = 11.84DD149 pKa = 3.75KK150 pKa = 11.6YY151 pKa = 10.36EE152 pKa = 4.72RR153 pKa = 11.84IHH155 pKa = 7.16NEE157 pKa = 3.34YY158 pKa = 10.2LARR161 pKa = 11.84TTTVFDD167 pKa = 4.51GRR169 pKa = 11.84LHH171 pKa = 7.42DD172 pKa = 4.25KK173 pKa = 10.79NIWIYY178 pKa = 10.8GPPGSGKK185 pKa = 8.61STLARR190 pKa = 11.84SDD192 pKa = 3.45TPIDD196 pKa = 3.87HH197 pKa = 7.38IYY199 pKa = 10.48FKK201 pKa = 10.78PVDD204 pKa = 3.66KK205 pKa = 10.17WWDD208 pKa = 3.37GFDD211 pKa = 3.55PSKK214 pKa = 10.86HH215 pKa = 5.28NRR217 pKa = 11.84IVMDD221 pKa = 4.89DD222 pKa = 4.06YY223 pKa = 11.57PNEE226 pKa = 4.12ATGGAIFTQRR236 pKa = 11.84MKK238 pKa = 10.35IWGDD242 pKa = 2.94RR243 pKa = 11.84TTEE246 pKa = 3.76PRR248 pKa = 11.84PVKK251 pKa = 10.53GGTITVDD258 pKa = 3.2PNIPFIVTSNFPIDD272 pKa = 3.42EE273 pKa = 4.47CFSNDD278 pKa = 2.85VDD280 pKa = 3.75RR281 pKa = 11.84EE282 pKa = 4.28AIHH285 pKa = 7.16RR286 pKa = 11.84RR287 pKa = 11.84FQEE290 pKa = 3.78IYY292 pKa = 10.97LDD294 pKa = 3.97GDD296 pKa = 3.32KK297 pKa = 11.37SKK299 pKa = 11.27LDD301 pKa = 3.21RR302 pKa = 11.84FMHH305 pKa = 6.42LKK307 pKa = 10.61NLINPEE313 pKa = 4.48DD314 pKa = 4.63PLDD317 pKa = 4.58CDD319 pKa = 4.6DD320 pKa = 6.25SIVSDD325 pKa = 3.69
Molecular weight: 38.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8E3Y3|G8E3Y3_9CIRC Uncharacterized protein OS=Po-Circo-like virus 41 OX=1105385 PE=4 SV=1
MM1 pKa = 7.61FGICNRR7 pKa = 11.84GSIIFLMVYY16 pKa = 9.1RR17 pKa = 11.84RR18 pKa = 11.84SYY20 pKa = 8.99RR21 pKa = 11.84RR22 pKa = 11.84SYY24 pKa = 9.49SRR26 pKa = 11.84SYY28 pKa = 10.43SRR30 pKa = 11.84PVQPVRR36 pKa = 11.84YY37 pKa = 9.88SNEE40 pKa = 3.99TYY42 pKa = 10.95NVTFQYY48 pKa = 10.8QWTATPTSATVTMIPASEE66 pKa = 4.12VQGLRR71 pKa = 11.84KK72 pKa = 8.21TKK74 pKa = 10.68NFTLNISQSPTTNAQGVVQSTSSFIYY100 pKa = 10.34ALVYY104 pKa = 10.58VPAGTVAGAITIGNGNAAASLYY126 pKa = 10.35EE127 pKa = 4.05PNQNVICSGVCDD139 pKa = 3.84SSSGQLRR146 pKa = 11.84ISSRR150 pKa = 11.84LARR153 pKa = 11.84NLNSGDD159 pKa = 3.49SVALVLRR166 pKa = 11.84PTYY169 pKa = 8.1TTGTEE174 pKa = 4.24GNITRR179 pKa = 11.84ASVTLNFAIAYY190 pKa = 8.25
MM1 pKa = 7.61FGICNRR7 pKa = 11.84GSIIFLMVYY16 pKa = 9.1RR17 pKa = 11.84RR18 pKa = 11.84SYY20 pKa = 8.99RR21 pKa = 11.84RR22 pKa = 11.84SYY24 pKa = 9.49SRR26 pKa = 11.84SYY28 pKa = 10.43SRR30 pKa = 11.84PVQPVRR36 pKa = 11.84YY37 pKa = 9.88SNEE40 pKa = 3.99TYY42 pKa = 10.95NVTFQYY48 pKa = 10.8QWTATPTSATVTMIPASEE66 pKa = 4.12VQGLRR71 pKa = 11.84KK72 pKa = 8.21TKK74 pKa = 10.68NFTLNISQSPTTNAQGVVQSTSSFIYY100 pKa = 10.34ALVYY104 pKa = 10.58VPAGTVAGAITIGNGNAAASLYY126 pKa = 10.35EE127 pKa = 4.05PNQNVICSGVCDD139 pKa = 3.84SSSGQLRR146 pKa = 11.84ISSRR150 pKa = 11.84LARR153 pKa = 11.84NLNSGDD159 pKa = 3.49SVALVLRR166 pKa = 11.84PTYY169 pKa = 8.1TTGTEE174 pKa = 4.24GNITRR179 pKa = 11.84ASVTLNFAIAYY190 pKa = 8.25
Molecular weight: 20.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
706 |
190 |
325 |
235.3 |
26.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.674 ± 1.343 | 1.7 ± 0.145 |
5.807 ± 2.37 | 4.958 ± 1.518 |
4.958 ± 0.798 | 6.799 ± 0.879 |
2.125 ± 1.011 | 6.799 ± 0.209 |
3.966 ± 1.08 | 5.524 ± 0.808 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.125 ± 0.394 | 6.799 ± 0.591 |
5.524 ± 0.47 | 3.399 ± 0.614 |
5.807 ± 1.533 | 8.782 ± 2.369 |
8.074 ± 0.982 | 5.949 ± 1.096 |
1.275 ± 0.733 | 4.958 ± 1.557 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
