
Plasmodium ovale (malaria parasite P. ovale)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Plasmodium)
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6625 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
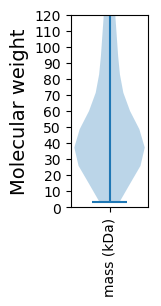
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1D3TGC1|A0A1D3TGC1_9APIC Transcription elongation factor SPT4 putative OS=Plasmodium ovale OX=36330 GN=SPT4 PE=3 SV=1
MM1 pKa = 6.48NTHH4 pKa = 6.75VILSTEE10 pKa = 3.95EE11 pKa = 3.84GGIVYY16 pKa = 10.08QATDD20 pKa = 3.42SRR22 pKa = 11.84DD23 pKa = 3.59LTDD26 pKa = 3.42MSEE29 pKa = 4.06PTDD32 pKa = 3.88LGEE35 pKa = 4.55SADD38 pKa = 4.08LGEE41 pKa = 4.46LTDD44 pKa = 3.57VGEE47 pKa = 4.24VTEE50 pKa = 4.15VGEE53 pKa = 4.1MTEE56 pKa = 3.74ASEE59 pKa = 4.32VIEE62 pKa = 4.51VGDD65 pKa = 3.58ATEE68 pKa = 4.05PTEE71 pKa = 4.05VGDD74 pKa = 3.58IGEE77 pKa = 3.98IAYY80 pKa = 10.15ASEE83 pKa = 3.94IGHH86 pKa = 5.95VGEE89 pKa = 5.07IGHH92 pKa = 6.81AGEE95 pKa = 4.81IGHH98 pKa = 6.92AGEE101 pKa = 4.81IGHH104 pKa = 6.95AGEE107 pKa = 4.08IGYY110 pKa = 10.53VGDD113 pKa = 3.23IAYY116 pKa = 9.94AHH118 pKa = 7.38DD119 pKa = 3.51IAYY122 pKa = 10.49VDD124 pKa = 5.24DD125 pKa = 4.08IAYY128 pKa = 10.41ADD130 pKa = 5.4DD131 pKa = 3.89IAHH134 pKa = 7.28ADD136 pKa = 4.47DD137 pKa = 4.39IAHH140 pKa = 7.27ADD142 pKa = 4.11DD143 pKa = 3.72IAYY146 pKa = 10.75SGDD149 pKa = 3.1ISNADD154 pKa = 3.81EE155 pKa = 3.84IAYY158 pKa = 8.26TSNVGYY164 pKa = 10.07ADD166 pKa = 3.34NMGYY170 pKa = 8.22TEE172 pKa = 4.24NLSYY176 pKa = 10.11TNNMDD181 pKa = 4.11HH182 pKa = 7.28IGDD185 pKa = 3.43EE186 pKa = 4.14RR187 pKa = 11.84YY188 pKa = 8.19YY189 pKa = 10.47TFNVDD194 pKa = 3.16YY195 pKa = 10.66TGDD198 pKa = 3.07QRR200 pKa = 11.84YY201 pKa = 6.25TVNVDD206 pKa = 3.12YY207 pKa = 11.08TGDD210 pKa = 3.52TRR212 pKa = 11.84HH213 pKa = 5.88VRR215 pKa = 11.84DD216 pKa = 3.42TRR218 pKa = 11.84YY219 pKa = 8.73TGDD222 pKa = 3.23TRR224 pKa = 11.84CTVNVGYY231 pKa = 8.58TRR233 pKa = 11.84NAGYY237 pKa = 8.66TGDD240 pKa = 3.55EE241 pKa = 4.9GYY243 pKa = 9.09TINVGYY249 pKa = 10.65VGDD252 pKa = 3.86AGYY255 pKa = 8.11TVNVGYY261 pKa = 10.51RR262 pKa = 11.84GDD264 pKa = 3.16ARR266 pKa = 11.84YY267 pKa = 7.33TVNVDD272 pKa = 3.17YY273 pKa = 10.55TVDD276 pKa = 3.53ADD278 pKa = 3.7HH279 pKa = 7.04TGGARR284 pKa = 11.84YY285 pKa = 7.25TLNVGYY291 pKa = 8.58TGDD294 pKa = 3.26GGYY297 pKa = 8.35TRR299 pKa = 11.84NSDD302 pKa = 3.26YY303 pKa = 10.83RR304 pKa = 11.84RR305 pKa = 11.84NSGYY309 pKa = 8.7TRR311 pKa = 11.84NARR314 pKa = 11.84YY315 pKa = 10.18SGDD318 pKa = 3.44VRR320 pKa = 11.84EE321 pKa = 4.41MATLTTASDD330 pKa = 4.31WIEE333 pKa = 3.84MADD336 pKa = 3.63VVNSGGMTEE345 pKa = 3.87MGEE348 pKa = 4.18VAEE351 pKa = 4.4EE352 pKa = 4.46TEE354 pKa = 4.27GTDD357 pKa = 3.55VIEE360 pKa = 4.64SNHH363 pKa = 4.86VAEE366 pKa = 4.65VTDD369 pKa = 3.79VTQLSDD375 pKa = 3.25TTYY378 pKa = 9.48DD379 pKa = 3.27TDD381 pKa = 3.48VTEE384 pKa = 5.2VIDD387 pKa = 3.81ATRR390 pKa = 11.84LTHH393 pKa = 5.85EE394 pKa = 4.45TDD396 pKa = 3.2VTQLSDD402 pKa = 3.66DD403 pKa = 3.89TDD405 pKa = 3.71VTDD408 pKa = 3.43VTEE411 pKa = 5.1VIDD414 pKa = 3.72ATQLSDD420 pKa = 3.26ATEE423 pKa = 4.09ATQFSDD429 pKa = 4.31VIEE432 pKa = 4.48EE433 pKa = 4.32NPSHH437 pKa = 7.32FSLL440 pKa = 5.2
MM1 pKa = 6.48NTHH4 pKa = 6.75VILSTEE10 pKa = 3.95EE11 pKa = 3.84GGIVYY16 pKa = 10.08QATDD20 pKa = 3.42SRR22 pKa = 11.84DD23 pKa = 3.59LTDD26 pKa = 3.42MSEE29 pKa = 4.06PTDD32 pKa = 3.88LGEE35 pKa = 4.55SADD38 pKa = 4.08LGEE41 pKa = 4.46LTDD44 pKa = 3.57VGEE47 pKa = 4.24VTEE50 pKa = 4.15VGEE53 pKa = 4.1MTEE56 pKa = 3.74ASEE59 pKa = 4.32VIEE62 pKa = 4.51VGDD65 pKa = 3.58ATEE68 pKa = 4.05PTEE71 pKa = 4.05VGDD74 pKa = 3.58IGEE77 pKa = 3.98IAYY80 pKa = 10.15ASEE83 pKa = 3.94IGHH86 pKa = 5.95VGEE89 pKa = 5.07IGHH92 pKa = 6.81AGEE95 pKa = 4.81IGHH98 pKa = 6.92AGEE101 pKa = 4.81IGHH104 pKa = 6.95AGEE107 pKa = 4.08IGYY110 pKa = 10.53VGDD113 pKa = 3.23IAYY116 pKa = 9.94AHH118 pKa = 7.38DD119 pKa = 3.51IAYY122 pKa = 10.49VDD124 pKa = 5.24DD125 pKa = 4.08IAYY128 pKa = 10.41ADD130 pKa = 5.4DD131 pKa = 3.89IAHH134 pKa = 7.28ADD136 pKa = 4.47DD137 pKa = 4.39IAHH140 pKa = 7.27ADD142 pKa = 4.11DD143 pKa = 3.72IAYY146 pKa = 10.75SGDD149 pKa = 3.1ISNADD154 pKa = 3.81EE155 pKa = 3.84IAYY158 pKa = 8.26TSNVGYY164 pKa = 10.07ADD166 pKa = 3.34NMGYY170 pKa = 8.22TEE172 pKa = 4.24NLSYY176 pKa = 10.11TNNMDD181 pKa = 4.11HH182 pKa = 7.28IGDD185 pKa = 3.43EE186 pKa = 4.14RR187 pKa = 11.84YY188 pKa = 8.19YY189 pKa = 10.47TFNVDD194 pKa = 3.16YY195 pKa = 10.66TGDD198 pKa = 3.07QRR200 pKa = 11.84YY201 pKa = 6.25TVNVDD206 pKa = 3.12YY207 pKa = 11.08TGDD210 pKa = 3.52TRR212 pKa = 11.84HH213 pKa = 5.88VRR215 pKa = 11.84DD216 pKa = 3.42TRR218 pKa = 11.84YY219 pKa = 8.73TGDD222 pKa = 3.23TRR224 pKa = 11.84CTVNVGYY231 pKa = 8.58TRR233 pKa = 11.84NAGYY237 pKa = 8.66TGDD240 pKa = 3.55EE241 pKa = 4.9GYY243 pKa = 9.09TINVGYY249 pKa = 10.65VGDD252 pKa = 3.86AGYY255 pKa = 8.11TVNVGYY261 pKa = 10.51RR262 pKa = 11.84GDD264 pKa = 3.16ARR266 pKa = 11.84YY267 pKa = 7.33TVNVDD272 pKa = 3.17YY273 pKa = 10.55TVDD276 pKa = 3.53ADD278 pKa = 3.7HH279 pKa = 7.04TGGARR284 pKa = 11.84YY285 pKa = 7.25TLNVGYY291 pKa = 8.58TGDD294 pKa = 3.26GGYY297 pKa = 8.35TRR299 pKa = 11.84NSDD302 pKa = 3.26YY303 pKa = 10.83RR304 pKa = 11.84RR305 pKa = 11.84NSGYY309 pKa = 8.7TRR311 pKa = 11.84NARR314 pKa = 11.84YY315 pKa = 10.18SGDD318 pKa = 3.44VRR320 pKa = 11.84EE321 pKa = 4.41MATLTTASDD330 pKa = 4.31WIEE333 pKa = 3.84MADD336 pKa = 3.63VVNSGGMTEE345 pKa = 3.87MGEE348 pKa = 4.18VAEE351 pKa = 4.4EE352 pKa = 4.46TEE354 pKa = 4.27GTDD357 pKa = 3.55VIEE360 pKa = 4.64SNHH363 pKa = 4.86VAEE366 pKa = 4.65VTDD369 pKa = 3.79VTQLSDD375 pKa = 3.25TTYY378 pKa = 9.48DD379 pKa = 3.27TDD381 pKa = 3.48VTEE384 pKa = 5.2VIDD387 pKa = 3.81ATRR390 pKa = 11.84LTHH393 pKa = 5.85EE394 pKa = 4.45TDD396 pKa = 3.2VTQLSDD402 pKa = 3.66DD403 pKa = 3.89TDD405 pKa = 3.71VTDD408 pKa = 3.43VTEE411 pKa = 5.1VIDD414 pKa = 3.72ATQLSDD420 pKa = 3.26ATEE423 pKa = 4.09ATQFSDD429 pKa = 4.31VIEE432 pKa = 4.48EE433 pKa = 4.32NPSHH437 pKa = 7.32FSLL440 pKa = 5.2
Molecular weight: 47.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D3TIQ4|A0A1D3TIQ4_9APIC Uncharacterized protein OS=Plasmodium ovale OX=36330 GN=PocGH01_11012700 PE=4 SV=1
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4221578 |
29 |
9915 |
637.2 |
73.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.14 ± 0.023 | 2.078 ± 0.013 |
5.733 ± 0.023 | 7.922 ± 0.035 |
4.675 ± 0.025 | 4.739 ± 0.033 |
2.396 ± 0.012 | 7.586 ± 0.03 |
10.497 ± 0.038 | 7.939 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.125 ± 0.009 | 9.423 ± 0.041 |
2.423 ± 0.019 | 2.658 ± 0.015 |
4.019 ± 0.021 | 7.784 ± 0.03 |
4.534 ± 0.017 | 4.713 ± 0.019 |
0.623 ± 0.007 | 4.988 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
