
Soil-borne cereal mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Furovirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
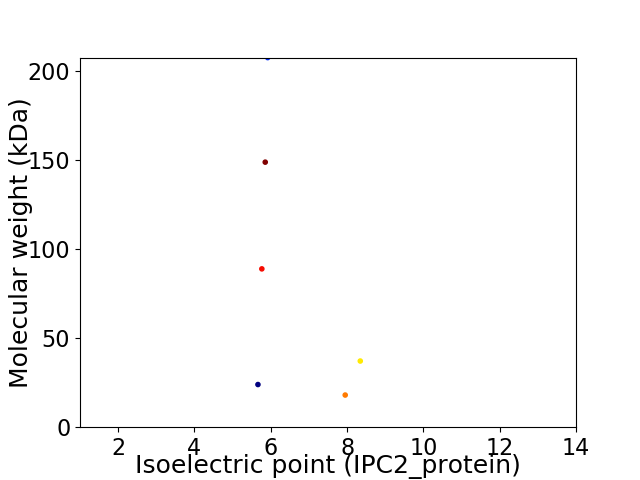
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q9QCE7|Q9QCE7_9VIRU Putative movement protein OS=Soil-borne cereal mosaic virus OX=100887 GN=ORF3 PE=4 SV=1
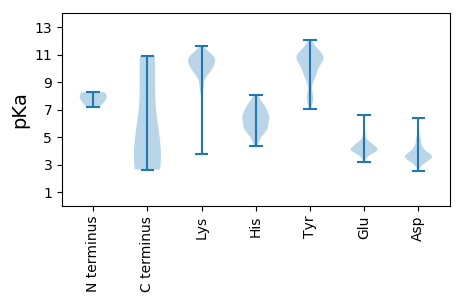
MM1 pKa = 8.27DD2 pKa = 5.47KK3 pKa = 10.11LTRR6 pKa = 11.84LKK8 pKa = 11.35GEE10 pKa = 4.17VDD12 pKa = 3.63RR13 pKa = 11.84SLQKK17 pKa = 10.96AADD20 pKa = 3.68KK21 pKa = 9.92VTAEE25 pKa = 4.39STEE28 pKa = 4.14SLQSKK33 pKa = 8.96VVVQLPQMAVKK44 pKa = 10.55NGYY47 pKa = 7.5TGYY50 pKa = 10.76NKK52 pKa = 9.98EE53 pKa = 4.34LNLMASIHH61 pKa = 5.47PFIRR65 pKa = 11.84LGTLISQIEE74 pKa = 4.46GWQATRR80 pKa = 11.84ASILTHH86 pKa = 6.51LGVMLNGVSKK96 pKa = 10.86LGEE99 pKa = 3.82RR100 pKa = 11.84NFFSRR105 pKa = 11.84QKK107 pKa = 10.75RR108 pKa = 11.84FGTHH112 pKa = 4.74TQDD115 pKa = 3.63GDD117 pKa = 4.7EE118 pKa = 4.48IFCDD122 pKa = 3.66LGGEE126 pKa = 3.95AVMQIITRR134 pKa = 11.84LTVSLQSAKK143 pKa = 11.07GEE145 pKa = 4.16GSQTRR150 pKa = 11.84NAMIGTTPTTNQFEE164 pKa = 4.61GEE166 pKa = 4.22EE167 pKa = 3.99QGQTDD172 pKa = 3.5QTLAISNALAEE183 pKa = 4.56FMTFIHH189 pKa = 6.29TKK191 pKa = 10.66DD192 pKa = 3.26FTTNEE197 pKa = 5.1CYY199 pKa = 9.71TQDD202 pKa = 3.26SFEE205 pKa = 4.94AKK207 pKa = 10.26FNLKK211 pKa = 9.68WEE213 pKa = 4.57GTSS216 pKa = 3.25
MM1 pKa = 8.27DD2 pKa = 5.47KK3 pKa = 10.11LTRR6 pKa = 11.84LKK8 pKa = 11.35GEE10 pKa = 4.17VDD12 pKa = 3.63RR13 pKa = 11.84SLQKK17 pKa = 10.96AADD20 pKa = 3.68KK21 pKa = 9.92VTAEE25 pKa = 4.39STEE28 pKa = 4.14SLQSKK33 pKa = 8.96VVVQLPQMAVKK44 pKa = 10.55NGYY47 pKa = 7.5TGYY50 pKa = 10.76NKK52 pKa = 9.98EE53 pKa = 4.34LNLMASIHH61 pKa = 5.47PFIRR65 pKa = 11.84LGTLISQIEE74 pKa = 4.46GWQATRR80 pKa = 11.84ASILTHH86 pKa = 6.51LGVMLNGVSKK96 pKa = 10.86LGEE99 pKa = 3.82RR100 pKa = 11.84NFFSRR105 pKa = 11.84QKK107 pKa = 10.75RR108 pKa = 11.84FGTHH112 pKa = 4.74TQDD115 pKa = 3.63GDD117 pKa = 4.7EE118 pKa = 4.48IFCDD122 pKa = 3.66LGGEE126 pKa = 3.95AVMQIITRR134 pKa = 11.84LTVSLQSAKK143 pKa = 11.07GEE145 pKa = 4.16GSQTRR150 pKa = 11.84NAMIGTTPTTNQFEE164 pKa = 4.61GEE166 pKa = 4.22EE167 pKa = 3.99QGQTDD172 pKa = 3.5QTLAISNALAEE183 pKa = 4.56FMTFIHH189 pKa = 6.29TKK191 pKa = 10.66DD192 pKa = 3.26FTTNEE197 pKa = 5.1CYY199 pKa = 9.71TQDD202 pKa = 3.26SFEE205 pKa = 4.94AKK207 pKa = 10.26FNLKK211 pKa = 9.68WEE213 pKa = 4.57GTSS216 pKa = 3.25
Molecular weight: 23.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QCE8|Q9QCE8_9VIRU Replicase OS=Soil-borne cereal mosaic virus OX=100887 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.51GSKK4 pKa = 9.97DD5 pKa = 3.5VKK7 pKa = 10.59GDD9 pKa = 3.58SFEE12 pKa = 4.35SVYY15 pKa = 11.47NKK17 pKa = 10.12ILDD20 pKa = 4.02LQTEE24 pKa = 4.54GAVGADD30 pKa = 3.11SLKK33 pKa = 10.16TNRR36 pKa = 11.84QRR38 pKa = 11.84SFNIEE43 pKa = 3.42NKK45 pKa = 9.46YY46 pKa = 9.71VEE48 pKa = 4.41KK49 pKa = 10.92ALVQQDD55 pKa = 3.11IATKK59 pKa = 9.51MRR61 pKa = 11.84EE62 pKa = 3.92GWTTWTKK69 pKa = 8.77TNKK72 pKa = 10.23EE73 pKa = 3.79EE74 pKa = 4.04GTPYY78 pKa = 10.91NMSYY82 pKa = 10.83SCVLLNVIPTVPRR95 pKa = 11.84GYY97 pKa = 10.36SGTVEE102 pKa = 3.61ISLIDD107 pKa = 3.63SGLSPLEE114 pKa = 3.86NVIPDD119 pKa = 3.58QTQMMEE125 pKa = 4.71LGNGPHH131 pKa = 5.95VMCFFMHH138 pKa = 6.43YY139 pKa = 10.1GIPLNDD145 pKa = 3.69EE146 pKa = 3.75GRR148 pKa = 11.84VIKK151 pKa = 10.72LSFKK155 pKa = 10.5IEE157 pKa = 3.67AEE159 pKa = 4.09MASKK163 pKa = 10.55KK164 pKa = 9.47MSVMNVYY171 pKa = 10.5SYY173 pKa = 7.61WTQKK177 pKa = 9.95QGHH180 pKa = 6.52LSVYY184 pKa = 9.15TEE186 pKa = 4.3PQRR189 pKa = 11.84STCSKK194 pKa = 10.8LLLGYY199 pKa = 10.47DD200 pKa = 3.5KK201 pKa = 10.98SLKK204 pKa = 10.61LKK206 pKa = 9.66TRR208 pKa = 11.84DD209 pKa = 3.25DD210 pKa = 3.34VRR212 pKa = 11.84RR213 pKa = 11.84FVPRR217 pKa = 11.84SLATNNLEE225 pKa = 3.93QSVPKK230 pKa = 9.86MLPNSINILKK240 pKa = 10.53EE241 pKa = 3.7NVPLFKK247 pKa = 10.61KK248 pKa = 10.67EE249 pKa = 4.1SVLDD253 pKa = 3.5LTEE256 pKa = 4.21QEE258 pKa = 4.26RR259 pKa = 11.84EE260 pKa = 4.09KK261 pKa = 10.86QKK263 pKa = 11.07KK264 pKa = 10.14LEE266 pKa = 4.07TLRR269 pKa = 11.84SVNEE273 pKa = 3.57AQRR276 pKa = 11.84RR277 pKa = 11.84KK278 pKa = 9.71SAQEE282 pKa = 3.55MQKK285 pKa = 10.7RR286 pKa = 11.84NDD288 pKa = 3.52DD289 pKa = 4.4LIRR292 pKa = 11.84ANVQKK297 pKa = 10.74QKK299 pKa = 11.03AKK301 pKa = 10.36QSEE304 pKa = 3.79IDD306 pKa = 3.63EE307 pKa = 4.33NRR309 pKa = 11.84KK310 pKa = 8.43NHH312 pKa = 6.09YY313 pKa = 9.6DD314 pKa = 3.29VSGVKK319 pKa = 10.27FGSVGSDD326 pKa = 2.64
MM1 pKa = 7.51GSKK4 pKa = 9.97DD5 pKa = 3.5VKK7 pKa = 10.59GDD9 pKa = 3.58SFEE12 pKa = 4.35SVYY15 pKa = 11.47NKK17 pKa = 10.12ILDD20 pKa = 4.02LQTEE24 pKa = 4.54GAVGADD30 pKa = 3.11SLKK33 pKa = 10.16TNRR36 pKa = 11.84QRR38 pKa = 11.84SFNIEE43 pKa = 3.42NKK45 pKa = 9.46YY46 pKa = 9.71VEE48 pKa = 4.41KK49 pKa = 10.92ALVQQDD55 pKa = 3.11IATKK59 pKa = 9.51MRR61 pKa = 11.84EE62 pKa = 3.92GWTTWTKK69 pKa = 8.77TNKK72 pKa = 10.23EE73 pKa = 3.79EE74 pKa = 4.04GTPYY78 pKa = 10.91NMSYY82 pKa = 10.83SCVLLNVIPTVPRR95 pKa = 11.84GYY97 pKa = 10.36SGTVEE102 pKa = 3.61ISLIDD107 pKa = 3.63SGLSPLEE114 pKa = 3.86NVIPDD119 pKa = 3.58QTQMMEE125 pKa = 4.71LGNGPHH131 pKa = 5.95VMCFFMHH138 pKa = 6.43YY139 pKa = 10.1GIPLNDD145 pKa = 3.69EE146 pKa = 3.75GRR148 pKa = 11.84VIKK151 pKa = 10.72LSFKK155 pKa = 10.5IEE157 pKa = 3.67AEE159 pKa = 4.09MASKK163 pKa = 10.55KK164 pKa = 9.47MSVMNVYY171 pKa = 10.5SYY173 pKa = 7.61WTQKK177 pKa = 9.95QGHH180 pKa = 6.52LSVYY184 pKa = 9.15TEE186 pKa = 4.3PQRR189 pKa = 11.84STCSKK194 pKa = 10.8LLLGYY199 pKa = 10.47DD200 pKa = 3.5KK201 pKa = 10.98SLKK204 pKa = 10.61LKK206 pKa = 9.66TRR208 pKa = 11.84DD209 pKa = 3.25DD210 pKa = 3.34VRR212 pKa = 11.84RR213 pKa = 11.84FVPRR217 pKa = 11.84SLATNNLEE225 pKa = 3.93QSVPKK230 pKa = 9.86MLPNSINILKK240 pKa = 10.53EE241 pKa = 3.7NVPLFKK247 pKa = 10.61KK248 pKa = 10.67EE249 pKa = 4.1SVLDD253 pKa = 3.5LTEE256 pKa = 4.21QEE258 pKa = 4.26RR259 pKa = 11.84EE260 pKa = 4.09KK261 pKa = 10.86QKK263 pKa = 11.07KK264 pKa = 10.14LEE266 pKa = 4.07TLRR269 pKa = 11.84SVNEE273 pKa = 3.57AQRR276 pKa = 11.84RR277 pKa = 11.84KK278 pKa = 9.71SAQEE282 pKa = 3.55MQKK285 pKa = 10.7RR286 pKa = 11.84NDD288 pKa = 3.52DD289 pKa = 4.4LIRR292 pKa = 11.84ANVQKK297 pKa = 10.74QKK299 pKa = 11.03AKK301 pKa = 10.36QSEE304 pKa = 3.79IDD306 pKa = 3.63EE307 pKa = 4.33NRR309 pKa = 11.84KK310 pKa = 8.43NHH312 pKa = 6.09YY313 pKa = 9.6DD314 pKa = 3.29VSGVKK319 pKa = 10.27FGSVGSDD326 pKa = 2.64
Molecular weight: 37.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4625 |
165 |
1816 |
770.8 |
87.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.681 ± 0.475 | 1.751 ± 0.283 |
6.724 ± 0.552 | 7.157 ± 0.207 |
4.13 ± 0.349 | 5.514 ± 0.561 |
2.097 ± 0.173 | 4.822 ± 0.216 |
7.222 ± 0.687 | 8.951 ± 0.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.789 ± 0.17 | 4.411 ± 0.342 |
2.789 ± 0.225 | 3.308 ± 0.614 |
6.054 ± 0.343 | 6.876 ± 0.431 |
6.811 ± 0.465 | 7.286 ± 0.36 |
1.189 ± 0.086 | 3.395 ± 0.366 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
