
Phlebovirus GGP-2011a
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; unclassified Phlebovirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
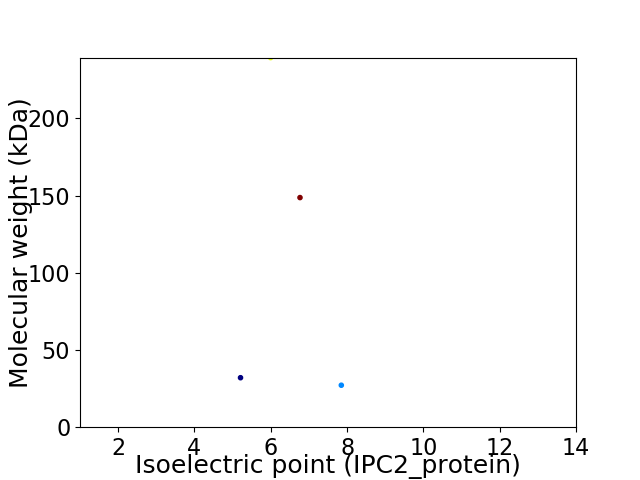
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1T371|I1T371_9VIRU Nucleoprotein OS=Phlebovirus GGP-2011a OX=1048857 PE=3 SV=1
MM1 pKa = 7.11NRR3 pKa = 11.84FMRR6 pKa = 11.84DD7 pKa = 3.02MPLVGTTGLSLNRR20 pKa = 11.84IQVTYY25 pKa = 10.75IPFNKK30 pKa = 8.47TWNSPVSTYY39 pKa = 11.51DD40 pKa = 3.19VMEE43 pKa = 4.91FPVHH47 pKa = 6.91NYY49 pKa = 7.1TQANEE54 pKa = 3.96IRR56 pKa = 11.84DD57 pKa = 3.99NLQSFYY63 pKa = 11.08RR64 pKa = 11.84LNRR67 pKa = 11.84IPARR71 pKa = 11.84WGPGFPSIRR80 pKa = 11.84NEE82 pKa = 3.54SDD84 pKa = 3.03IEE86 pKa = 4.16HH87 pKa = 7.05LSLIHH92 pKa = 6.34EE93 pKa = 4.64LSKK96 pKa = 10.25IDD98 pKa = 3.83EE99 pKa = 4.38IEE101 pKa = 3.96IVRR104 pKa = 11.84HH105 pKa = 5.31NEE107 pKa = 3.78PNLKK111 pKa = 9.96RR112 pKa = 11.84ALYY115 pKa = 9.56WPFNYY120 pKa = 9.85PSMSFIRR127 pKa = 11.84HH128 pKa = 5.93ASRR131 pKa = 11.84VDD133 pKa = 3.58EE134 pKa = 4.41EE135 pKa = 4.71KK136 pKa = 10.51PWLFKK141 pKa = 11.55NNVATDD147 pKa = 3.4ILRR150 pKa = 11.84ASKK153 pKa = 9.28STVLDD158 pKa = 3.7LAIVNLHH165 pKa = 6.13RR166 pKa = 11.84EE167 pKa = 4.33VLRR170 pKa = 11.84EE171 pKa = 3.98AARR174 pKa = 11.84QSLPEE179 pKa = 4.03GKK181 pKa = 10.4FPGKK185 pKa = 10.19DD186 pKa = 2.65ILMEE190 pKa = 4.22IASLQCRR197 pKa = 11.84RR198 pKa = 11.84MLSAIDD204 pKa = 3.9SDD206 pKa = 4.25SEE208 pKa = 4.21YY209 pKa = 10.81DD210 pKa = 3.96GPHH213 pKa = 5.83SPIFNAVWDD222 pKa = 3.99YY223 pKa = 11.58RR224 pKa = 11.84EE225 pKa = 4.21SCLEE229 pKa = 3.97TVFFSDD235 pKa = 3.39WVPSVDD241 pKa = 5.47EE242 pKa = 4.14FDD244 pKa = 4.1CPIMHH249 pKa = 7.04MMRR252 pKa = 11.84NGLRR256 pKa = 11.84TWDD259 pKa = 3.96PEE261 pKa = 4.14DD262 pKa = 3.87TEE264 pKa = 4.82PEE266 pKa = 3.54VDD268 pKa = 3.15AASGLKK274 pKa = 10.66ANN276 pKa = 4.61
MM1 pKa = 7.11NRR3 pKa = 11.84FMRR6 pKa = 11.84DD7 pKa = 3.02MPLVGTTGLSLNRR20 pKa = 11.84IQVTYY25 pKa = 10.75IPFNKK30 pKa = 8.47TWNSPVSTYY39 pKa = 11.51DD40 pKa = 3.19VMEE43 pKa = 4.91FPVHH47 pKa = 6.91NYY49 pKa = 7.1TQANEE54 pKa = 3.96IRR56 pKa = 11.84DD57 pKa = 3.99NLQSFYY63 pKa = 11.08RR64 pKa = 11.84LNRR67 pKa = 11.84IPARR71 pKa = 11.84WGPGFPSIRR80 pKa = 11.84NEE82 pKa = 3.54SDD84 pKa = 3.03IEE86 pKa = 4.16HH87 pKa = 7.05LSLIHH92 pKa = 6.34EE93 pKa = 4.64LSKK96 pKa = 10.25IDD98 pKa = 3.83EE99 pKa = 4.38IEE101 pKa = 3.96IVRR104 pKa = 11.84HH105 pKa = 5.31NEE107 pKa = 3.78PNLKK111 pKa = 9.96RR112 pKa = 11.84ALYY115 pKa = 9.56WPFNYY120 pKa = 9.85PSMSFIRR127 pKa = 11.84HH128 pKa = 5.93ASRR131 pKa = 11.84VDD133 pKa = 3.58EE134 pKa = 4.41EE135 pKa = 4.71KK136 pKa = 10.51PWLFKK141 pKa = 11.55NNVATDD147 pKa = 3.4ILRR150 pKa = 11.84ASKK153 pKa = 9.28STVLDD158 pKa = 3.7LAIVNLHH165 pKa = 6.13RR166 pKa = 11.84EE167 pKa = 4.33VLRR170 pKa = 11.84EE171 pKa = 3.98AARR174 pKa = 11.84QSLPEE179 pKa = 4.03GKK181 pKa = 10.4FPGKK185 pKa = 10.19DD186 pKa = 2.65ILMEE190 pKa = 4.22IASLQCRR197 pKa = 11.84RR198 pKa = 11.84MLSAIDD204 pKa = 3.9SDD206 pKa = 4.25SEE208 pKa = 4.21YY209 pKa = 10.81DD210 pKa = 3.96GPHH213 pKa = 5.83SPIFNAVWDD222 pKa = 3.99YY223 pKa = 11.58RR224 pKa = 11.84EE225 pKa = 4.21SCLEE229 pKa = 3.97TVFFSDD235 pKa = 3.39WVPSVDD241 pKa = 5.47EE242 pKa = 4.14FDD244 pKa = 4.1CPIMHH249 pKa = 7.04MMRR252 pKa = 11.84NGLRR256 pKa = 11.84TWDD259 pKa = 3.96PEE261 pKa = 4.14DD262 pKa = 3.87TEE264 pKa = 4.82PEE266 pKa = 3.54VDD268 pKa = 3.15AASGLKK274 pKa = 10.66ANN276 pKa = 4.61
Molecular weight: 32.05 kDa
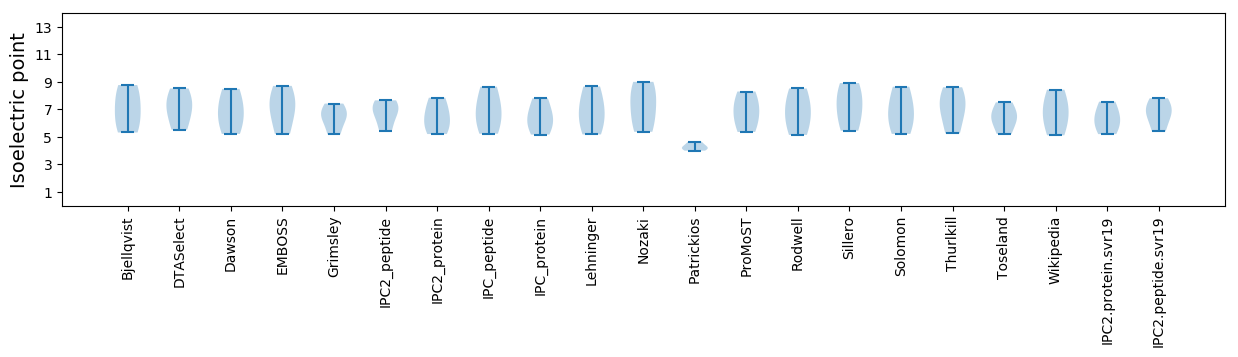
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1T371|I1T371_9VIRU Nucleoprotein OS=Phlebovirus GGP-2011a OX=1048857 PE=3 SV=1
MM1 pKa = 7.69SDD3 pKa = 3.5YY4 pKa = 11.17EE5 pKa = 4.26RR6 pKa = 11.84IAVEE10 pKa = 4.36FAADD14 pKa = 3.3IPSNEE19 pKa = 4.47VIQEE23 pKa = 3.94WVEE26 pKa = 3.67AFAYY30 pKa = 10.22QGFDD34 pKa = 2.98AAHH37 pKa = 5.18VLKK40 pKa = 10.75RR41 pKa = 11.84LIEE44 pKa = 4.42CGGNNWKK51 pKa = 10.22EE52 pKa = 3.95DD53 pKa = 3.49AKK55 pKa = 10.97KK56 pKa = 10.71LIVIAITRR64 pKa = 11.84GNKK67 pKa = 8.4PDD69 pKa = 4.19KK70 pKa = 10.82IMEE73 pKa = 4.34KK74 pKa = 9.73MSEE77 pKa = 3.97EE78 pKa = 3.9GKK80 pKa = 10.61KK81 pKa = 10.39AFKK84 pKa = 10.29VLKK87 pKa = 10.15DD88 pKa = 3.57RR89 pKa = 11.84YY90 pKa = 9.17RR91 pKa = 11.84LQGGNPGRR99 pKa = 11.84SDD101 pKa = 3.34LTLSRR106 pKa = 11.84IATALAGWTCQACVVVGEE124 pKa = 4.3YY125 pKa = 10.97LPVTIGTMRR134 pKa = 11.84SISDD138 pKa = 3.76NYY140 pKa = 8.69PGAMMHH146 pKa = 6.69PCFAGLIDD154 pKa = 4.33RR155 pKa = 11.84SLPEE159 pKa = 3.96DD160 pKa = 3.52TKK162 pKa = 11.43LVIINAFCLFMVHH175 pKa = 5.56FTKK178 pKa = 10.79VINPRR183 pKa = 11.84MRR185 pKa = 11.84SSPVSEE191 pKa = 4.12VVNSFRR197 pKa = 11.84QPMLAAINSNFLNSDD212 pKa = 2.98QRR214 pKa = 11.84KK215 pKa = 9.16GFLKK219 pKa = 10.83SLGILDD225 pKa = 5.53ANLQPTAPVRR235 pKa = 11.84AAARR239 pKa = 11.84AYY241 pKa = 9.97EE242 pKa = 4.06LLAA245 pKa = 5.59
MM1 pKa = 7.69SDD3 pKa = 3.5YY4 pKa = 11.17EE5 pKa = 4.26RR6 pKa = 11.84IAVEE10 pKa = 4.36FAADD14 pKa = 3.3IPSNEE19 pKa = 4.47VIQEE23 pKa = 3.94WVEE26 pKa = 3.67AFAYY30 pKa = 10.22QGFDD34 pKa = 2.98AAHH37 pKa = 5.18VLKK40 pKa = 10.75RR41 pKa = 11.84LIEE44 pKa = 4.42CGGNNWKK51 pKa = 10.22EE52 pKa = 3.95DD53 pKa = 3.49AKK55 pKa = 10.97KK56 pKa = 10.71LIVIAITRR64 pKa = 11.84GNKK67 pKa = 8.4PDD69 pKa = 4.19KK70 pKa = 10.82IMEE73 pKa = 4.34KK74 pKa = 9.73MSEE77 pKa = 3.97EE78 pKa = 3.9GKK80 pKa = 10.61KK81 pKa = 10.39AFKK84 pKa = 10.29VLKK87 pKa = 10.15DD88 pKa = 3.57RR89 pKa = 11.84YY90 pKa = 9.17RR91 pKa = 11.84LQGGNPGRR99 pKa = 11.84SDD101 pKa = 3.34LTLSRR106 pKa = 11.84IATALAGWTCQACVVVGEE124 pKa = 4.3YY125 pKa = 10.97LPVTIGTMRR134 pKa = 11.84SISDD138 pKa = 3.76NYY140 pKa = 8.69PGAMMHH146 pKa = 6.69PCFAGLIDD154 pKa = 4.33RR155 pKa = 11.84SLPEE159 pKa = 3.96DD160 pKa = 3.52TKK162 pKa = 11.43LVIINAFCLFMVHH175 pKa = 5.56FTKK178 pKa = 10.79VINPRR183 pKa = 11.84MRR185 pKa = 11.84SSPVSEE191 pKa = 4.12VVNSFRR197 pKa = 11.84QPMLAAINSNFLNSDD212 pKa = 2.98QRR214 pKa = 11.84KK215 pKa = 9.16GFLKK219 pKa = 10.83SLGILDD225 pKa = 5.53ANLQPTAPVRR235 pKa = 11.84AAARR239 pKa = 11.84AYY241 pKa = 9.97EE242 pKa = 4.06LLAA245 pKa = 5.59
Molecular weight: 27.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3958 |
245 |
2098 |
989.5 |
111.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.533 ± 0.594 | 2.602 ± 0.684 |
5.533 ± 0.352 | 6.948 ± 0.47 |
4.396 ± 0.473 | 5.659 ± 0.359 |
2.602 ± 0.298 | 6.721 ± 0.108 |
6.139 ± 0.408 | 8.489 ± 0.292 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.234 ± 0.369 | 4.144 ± 0.534 |
4.295 ± 0.365 | 2.83 ± 0.128 |
5.76 ± 0.355 | 9.323 ± 0.465 |
5.129 ± 0.41 | 6.392 ± 0.442 |
1.364 ± 0.14 | 2.906 ± 0.105 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
