
Streptococcus pasteurianus (strain ATCC 43144 / JCM 5346 / CDC 1723-81)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus pasteurianus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
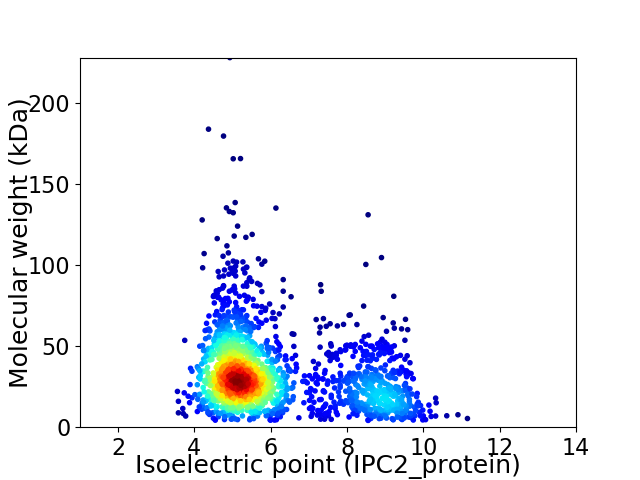
Virtual 2D-PAGE plot for 1864 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5X2E7|F5X2E7_STRPX Predicted membrane protein OS=Streptococcus pasteurianus (strain ATCC 43144 / JCM 5346 / CDC 1723-81) OX=981540 GN=SGPB_1420 PE=4 SV=1
MM1 pKa = 7.42PTRR4 pKa = 11.84NYY6 pKa = 10.48SRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.05RR11 pKa = 11.84QQKK14 pKa = 8.77SQIIALTGLVTAAVALVLILAKK36 pKa = 10.67LIFSGVTTLASDD48 pKa = 4.29GSLDD52 pKa = 4.09LSDD55 pKa = 4.43ANSYY59 pKa = 10.42DD60 pKa = 3.26VSEE63 pKa = 4.44IATVSTSDD71 pKa = 3.87AIIYY75 pKa = 9.48TDD77 pKa = 4.74DD78 pKa = 3.99GSSSKK83 pKa = 10.35ISEE86 pKa = 4.03NTAITISAYY95 pKa = 10.81YY96 pKa = 9.11YY97 pKa = 10.69DD98 pKa = 5.47AMLNDD103 pKa = 4.26EE104 pKa = 5.05DD105 pKa = 3.96TTLAQFSMNGDD116 pKa = 3.53TYY118 pKa = 11.5YY119 pKa = 10.33IDD121 pKa = 3.68TNDD124 pKa = 3.95ISLEE128 pKa = 3.79QDD130 pKa = 3.04NTINAYY136 pKa = 9.26IAEE139 pKa = 4.32TLGYY143 pKa = 9.41SHH145 pKa = 7.5ADD147 pKa = 2.73ITDD150 pKa = 4.54DD151 pKa = 3.25IKK153 pKa = 11.62SSFEE157 pKa = 3.4QAAYY161 pKa = 8.35KK162 pKa = 10.15TDD164 pKa = 3.57DD165 pKa = 3.89GKK167 pKa = 10.1PLGVIIHH174 pKa = 6.48DD175 pKa = 3.8TGVDD179 pKa = 3.13NSTIEE184 pKa = 4.3SEE186 pKa = 4.38VNYY189 pKa = 9.55MVQNYY194 pKa = 9.52DD195 pKa = 3.19KK196 pKa = 10.66EE197 pKa = 4.78GVFVHH202 pKa = 6.45SFIDD206 pKa = 3.05SDD208 pKa = 4.26TILRR212 pKa = 11.84IADD215 pKa = 3.7EE216 pKa = 4.95GYY218 pKa = 9.73KK219 pKa = 10.59AQGAGANANPYY230 pKa = 9.49YY231 pKa = 10.18IQFEE235 pKa = 4.5LTHH238 pKa = 6.78EE239 pKa = 4.54DD240 pKa = 3.54SQKK243 pKa = 11.04GFAEE247 pKa = 3.95QLANAAYY254 pKa = 7.84YY255 pKa = 7.92TAYY258 pKa = 9.85MLKK261 pKa = 10.29KK262 pKa = 9.59YY263 pKa = 10.0DD264 pKa = 3.8LPVTLGQEE272 pKa = 4.26DD273 pKa = 4.81GEE275 pKa = 4.67GTIWTHH281 pKa = 5.77EE282 pKa = 4.24MVSLYY287 pKa = 10.84LGGTDD292 pKa = 4.6HH293 pKa = 7.64VDD295 pKa = 3.12PTDD298 pKa = 3.34YY299 pKa = 8.5WTEE302 pKa = 3.73TANDD306 pKa = 3.92YY307 pKa = 11.19FGTDD311 pKa = 3.11YY312 pKa = 11.1DD313 pKa = 3.69IEE315 pKa = 4.76DD316 pKa = 3.9FVEE319 pKa = 4.96LVQAYY324 pKa = 10.1YY325 pKa = 10.88NALL328 pKa = 3.56
MM1 pKa = 7.42PTRR4 pKa = 11.84NYY6 pKa = 10.48SRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.05RR11 pKa = 11.84QQKK14 pKa = 8.77SQIIALTGLVTAAVALVLILAKK36 pKa = 10.67LIFSGVTTLASDD48 pKa = 4.29GSLDD52 pKa = 4.09LSDD55 pKa = 4.43ANSYY59 pKa = 10.42DD60 pKa = 3.26VSEE63 pKa = 4.44IATVSTSDD71 pKa = 3.87AIIYY75 pKa = 9.48TDD77 pKa = 4.74DD78 pKa = 3.99GSSSKK83 pKa = 10.35ISEE86 pKa = 4.03NTAITISAYY95 pKa = 10.81YY96 pKa = 9.11YY97 pKa = 10.69DD98 pKa = 5.47AMLNDD103 pKa = 4.26EE104 pKa = 5.05DD105 pKa = 3.96TTLAQFSMNGDD116 pKa = 3.53TYY118 pKa = 11.5YY119 pKa = 10.33IDD121 pKa = 3.68TNDD124 pKa = 3.95ISLEE128 pKa = 3.79QDD130 pKa = 3.04NTINAYY136 pKa = 9.26IAEE139 pKa = 4.32TLGYY143 pKa = 9.41SHH145 pKa = 7.5ADD147 pKa = 2.73ITDD150 pKa = 4.54DD151 pKa = 3.25IKK153 pKa = 11.62SSFEE157 pKa = 3.4QAAYY161 pKa = 8.35KK162 pKa = 10.15TDD164 pKa = 3.57DD165 pKa = 3.89GKK167 pKa = 10.1PLGVIIHH174 pKa = 6.48DD175 pKa = 3.8TGVDD179 pKa = 3.13NSTIEE184 pKa = 4.3SEE186 pKa = 4.38VNYY189 pKa = 9.55MVQNYY194 pKa = 9.52DD195 pKa = 3.19KK196 pKa = 10.66EE197 pKa = 4.78GVFVHH202 pKa = 6.45SFIDD206 pKa = 3.05SDD208 pKa = 4.26TILRR212 pKa = 11.84IADD215 pKa = 3.7EE216 pKa = 4.95GYY218 pKa = 9.73KK219 pKa = 10.59AQGAGANANPYY230 pKa = 9.49YY231 pKa = 10.18IQFEE235 pKa = 4.5LTHH238 pKa = 6.78EE239 pKa = 4.54DD240 pKa = 3.54SQKK243 pKa = 11.04GFAEE247 pKa = 3.95QLANAAYY254 pKa = 7.84YY255 pKa = 7.92TAYY258 pKa = 9.85MLKK261 pKa = 10.29KK262 pKa = 9.59YY263 pKa = 10.0DD264 pKa = 3.8LPVTLGQEE272 pKa = 4.26DD273 pKa = 4.81GEE275 pKa = 4.67GTIWTHH281 pKa = 5.77EE282 pKa = 4.24MVSLYY287 pKa = 10.84LGGTDD292 pKa = 4.6HH293 pKa = 7.64VDD295 pKa = 3.12PTDD298 pKa = 3.34YY299 pKa = 8.5WTEE302 pKa = 3.73TANDD306 pKa = 3.92YY307 pKa = 11.19FGTDD311 pKa = 3.11YY312 pKa = 11.1DD313 pKa = 3.69IEE315 pKa = 4.76DD316 pKa = 3.9FVEE319 pKa = 4.96LVQAYY324 pKa = 10.1YY325 pKa = 10.88NALL328 pKa = 3.56
Molecular weight: 36.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5X3R9|F5X3R9_STRPX 30S ribosomal protein S2 OS=Streptococcus pasteurianus (strain ATCC 43144 / JCM 5346 / CDC 1723-81) OX=981540 GN=rpsB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.67IRR11 pKa = 11.84RR12 pKa = 11.84QRR14 pKa = 11.84KK15 pKa = 7.52HH16 pKa = 5.2GFRR19 pKa = 11.84HH20 pKa = 6.39RR21 pKa = 11.84MSTKK25 pKa = 9.02NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.67IRR11 pKa = 11.84RR12 pKa = 11.84QRR14 pKa = 11.84KK15 pKa = 7.52HH16 pKa = 5.2GFRR19 pKa = 11.84HH20 pKa = 6.39RR21 pKa = 11.84MSTKK25 pKa = 9.02NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
569143 |
38 |
1998 |
305.3 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.662 ± 0.056 | 0.568 ± 0.013 |
5.954 ± 0.048 | 6.81 ± 0.057 |
4.604 ± 0.043 | 6.488 ± 0.056 |
1.876 ± 0.023 | 7.596 ± 0.051 |
6.717 ± 0.046 | 9.871 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.026 | 4.719 ± 0.039 |
3.138 ± 0.029 | 3.743 ± 0.036 |
3.819 ± 0.042 | 6.204 ± 0.051 |
5.914 ± 0.047 | 7.106 ± 0.05 |
0.857 ± 0.018 | 3.802 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
