
Clostridium sp. CAG:1024
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
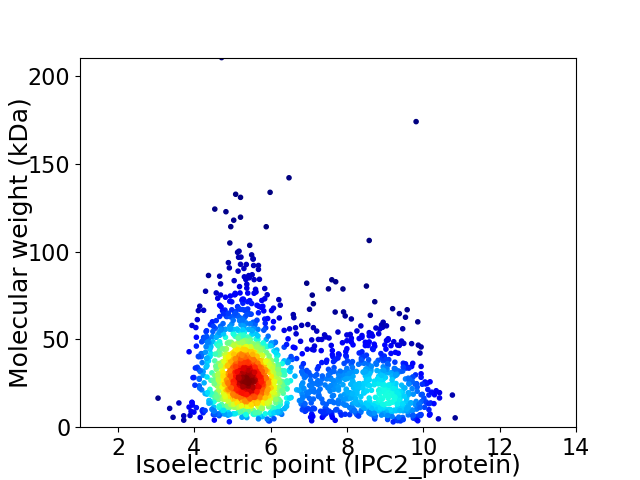
Virtual 2D-PAGE plot for 1915 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5AI68|R5AI68_9CLOT Phospho-N-acetylmuramoyl-pentapeptide-transferase OS=Clostridium sp. CAG:1024 OX=1262770 GN=mraY PE=3 SV=1
MM1 pKa = 7.78KK2 pKa = 9.33LTKK5 pKa = 9.61TVSLLAAVLLLVLSLTACQFGTLPSAGTAATPAEE39 pKa = 4.33DD40 pKa = 3.97TQDD43 pKa = 3.17PTMVYY48 pKa = 10.43LPDD51 pKa = 3.36TAAPVQGEE59 pKa = 4.51STDD62 pKa = 3.82TPDD65 pKa = 5.92DD66 pKa = 4.04DD67 pKa = 4.41AQRR70 pKa = 11.84LAAYY74 pKa = 10.02RR75 pKa = 11.84EE76 pKa = 4.35ALSSFYY82 pKa = 10.3STGRR86 pKa = 11.84FPDD89 pKa = 4.03GTDD92 pKa = 2.75SGYY95 pKa = 10.91SADD98 pKa = 3.83FGEE101 pKa = 4.35ITGNRR106 pKa = 11.84FAICDD111 pKa = 3.57VDD113 pKa = 4.66GDD115 pKa = 4.35GADD118 pKa = 3.61EE119 pKa = 5.72LILQFVTAPVAGQLEE134 pKa = 4.18IVYY137 pKa = 10.04RR138 pKa = 11.84YY139 pKa = 7.84DD140 pKa = 3.42TASGALVSEE149 pKa = 4.48LSEE152 pKa = 4.24YY153 pKa = 10.37PPLSYY158 pKa = 10.55GANGSVTALFSHH170 pKa = 5.98NQGLSGGDD178 pKa = 2.94FWPYY182 pKa = 9.85HH183 pKa = 6.47RR184 pKa = 11.84YY185 pKa = 9.95VFDD188 pKa = 4.65AAAQSYY194 pKa = 7.56TLEE197 pKa = 4.26ATVDD201 pKa = 2.89AWSRR205 pKa = 11.84AVSDD209 pKa = 3.68TDD211 pKa = 4.08FDD213 pKa = 4.84GNPYY217 pKa = 10.31PNDD220 pKa = 3.22IDD222 pKa = 4.73RR223 pKa = 11.84EE224 pKa = 4.37GAGIVYY230 pKa = 10.18LVTEE234 pKa = 4.34NGSTNTLSKK243 pKa = 10.64SDD245 pKa = 3.83YY246 pKa = 10.09EE247 pKa = 4.34SWLMSHH253 pKa = 7.47PSGAAILVPYY263 pKa = 10.08TEE265 pKa = 4.51LTEE268 pKa = 4.15SAIAAIGEE276 pKa = 4.34TAVNEE281 pKa = 3.94ISLSLFADD289 pKa = 3.68SAEE292 pKa = 4.11GLAALLDD299 pKa = 4.43NINARR304 pKa = 11.84VEE306 pKa = 4.1VGTAGSSLKK315 pKa = 9.57ATYY318 pKa = 10.21AAGQLLDD325 pKa = 3.19WCAATKK331 pKa = 10.64LSADD335 pKa = 3.98DD336 pKa = 3.63VRR338 pKa = 11.84AATLAWMVNLGNDD351 pKa = 3.37AQIAFAEE358 pKa = 4.28KK359 pKa = 10.22LALVNAACSSLRR371 pKa = 11.84TDD373 pKa = 3.56KK374 pKa = 11.38GADD377 pKa = 3.28LMSEE381 pKa = 4.06AGYY384 pKa = 9.8PSAAYY389 pKa = 8.47PWTTLSPEE397 pKa = 4.19TLSSIMEE404 pKa = 4.19AVGVEE409 pKa = 4.0
MM1 pKa = 7.78KK2 pKa = 9.33LTKK5 pKa = 9.61TVSLLAAVLLLVLSLTACQFGTLPSAGTAATPAEE39 pKa = 4.33DD40 pKa = 3.97TQDD43 pKa = 3.17PTMVYY48 pKa = 10.43LPDD51 pKa = 3.36TAAPVQGEE59 pKa = 4.51STDD62 pKa = 3.82TPDD65 pKa = 5.92DD66 pKa = 4.04DD67 pKa = 4.41AQRR70 pKa = 11.84LAAYY74 pKa = 10.02RR75 pKa = 11.84EE76 pKa = 4.35ALSSFYY82 pKa = 10.3STGRR86 pKa = 11.84FPDD89 pKa = 4.03GTDD92 pKa = 2.75SGYY95 pKa = 10.91SADD98 pKa = 3.83FGEE101 pKa = 4.35ITGNRR106 pKa = 11.84FAICDD111 pKa = 3.57VDD113 pKa = 4.66GDD115 pKa = 4.35GADD118 pKa = 3.61EE119 pKa = 5.72LILQFVTAPVAGQLEE134 pKa = 4.18IVYY137 pKa = 10.04RR138 pKa = 11.84YY139 pKa = 7.84DD140 pKa = 3.42TASGALVSEE149 pKa = 4.48LSEE152 pKa = 4.24YY153 pKa = 10.37PPLSYY158 pKa = 10.55GANGSVTALFSHH170 pKa = 5.98NQGLSGGDD178 pKa = 2.94FWPYY182 pKa = 9.85HH183 pKa = 6.47RR184 pKa = 11.84YY185 pKa = 9.95VFDD188 pKa = 4.65AAAQSYY194 pKa = 7.56TLEE197 pKa = 4.26ATVDD201 pKa = 2.89AWSRR205 pKa = 11.84AVSDD209 pKa = 3.68TDD211 pKa = 4.08FDD213 pKa = 4.84GNPYY217 pKa = 10.31PNDD220 pKa = 3.22IDD222 pKa = 4.73RR223 pKa = 11.84EE224 pKa = 4.37GAGIVYY230 pKa = 10.18LVTEE234 pKa = 4.34NGSTNTLSKK243 pKa = 10.64SDD245 pKa = 3.83YY246 pKa = 10.09EE247 pKa = 4.34SWLMSHH253 pKa = 7.47PSGAAILVPYY263 pKa = 10.08TEE265 pKa = 4.51LTEE268 pKa = 4.15SAIAAIGEE276 pKa = 4.34TAVNEE281 pKa = 3.94ISLSLFADD289 pKa = 3.68SAEE292 pKa = 4.11GLAALLDD299 pKa = 4.43NINARR304 pKa = 11.84VEE306 pKa = 4.1VGTAGSSLKK315 pKa = 9.57ATYY318 pKa = 10.21AAGQLLDD325 pKa = 3.19WCAATKK331 pKa = 10.64LSADD335 pKa = 3.98DD336 pKa = 3.63VRR338 pKa = 11.84AATLAWMVNLGNDD351 pKa = 3.37AQIAFAEE358 pKa = 4.28KK359 pKa = 10.22LALVNAACSSLRR371 pKa = 11.84TDD373 pKa = 3.56KK374 pKa = 11.38GADD377 pKa = 3.28LMSEE381 pKa = 4.06AGYY384 pKa = 9.8PSAAYY389 pKa = 8.47PWTTLSPEE397 pKa = 4.19TLSSIMEE404 pKa = 4.19AVGVEE409 pKa = 4.0
Molecular weight: 42.98 kDa
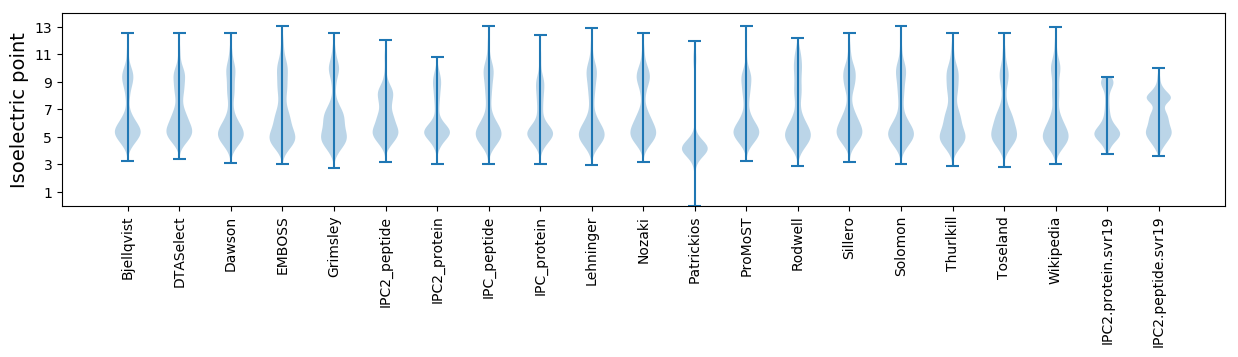
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5AE72|R5AE72_9CLOT Uncharacterized protein OS=Clostridium sp. CAG:1024 OX=1262770 GN=BN454_00847 PE=4 SV=1
MM1 pKa = 6.83TRR3 pKa = 11.84AAARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84AAIRR14 pKa = 11.84RR15 pKa = 11.84TITTSRR21 pKa = 11.84CPATDD26 pKa = 3.34RR27 pKa = 11.84VSCSCRR33 pKa = 11.84RR34 pKa = 11.84CKK36 pKa = 9.95WPALSRR42 pKa = 11.84PMRR45 pKa = 11.84TTAISWSPMWSIPSGMRR62 pKa = 11.84TARR65 pKa = 11.84TTTSCPRR72 pKa = 11.84PNRR75 pKa = 11.84ASGKK79 pKa = 8.49RR80 pKa = 11.84ARR82 pKa = 11.84SSRR85 pKa = 11.84ARR87 pKa = 11.84SIRR90 pKa = 11.84STRR93 pKa = 11.84PSQRR97 pKa = 11.84SATSARR103 pKa = 11.84RR104 pKa = 11.84SLCRR108 pKa = 11.84GSSTARR114 pKa = 11.84AISTPSATSSSARR127 pKa = 11.84RR128 pKa = 11.84VRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84SRR134 pKa = 11.84TTNPRR139 pKa = 11.84SSAGSSPGATAWSRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84GSAASCWRR164 pKa = 11.84SII166 pKa = 4.18
MM1 pKa = 6.83TRR3 pKa = 11.84AAARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84AAIRR14 pKa = 11.84RR15 pKa = 11.84TITTSRR21 pKa = 11.84CPATDD26 pKa = 3.34RR27 pKa = 11.84VSCSCRR33 pKa = 11.84RR34 pKa = 11.84CKK36 pKa = 9.95WPALSRR42 pKa = 11.84PMRR45 pKa = 11.84TTAISWSPMWSIPSGMRR62 pKa = 11.84TARR65 pKa = 11.84TTTSCPRR72 pKa = 11.84PNRR75 pKa = 11.84ASGKK79 pKa = 8.49RR80 pKa = 11.84ARR82 pKa = 11.84SSRR85 pKa = 11.84ARR87 pKa = 11.84SIRR90 pKa = 11.84STRR93 pKa = 11.84PSQRR97 pKa = 11.84SATSARR103 pKa = 11.84RR104 pKa = 11.84SLCRR108 pKa = 11.84GSSTARR114 pKa = 11.84AISTPSATSSSARR127 pKa = 11.84RR128 pKa = 11.84VRR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84SRR134 pKa = 11.84TTNPRR139 pKa = 11.84SSAGSSPGATAWSRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84GSAASCWRR164 pKa = 11.84SII166 pKa = 4.18
Molecular weight: 18.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
549254 |
29 |
1928 |
286.8 |
31.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.335 ± 0.071 | 1.728 ± 0.024 |
5.503 ± 0.048 | 6.662 ± 0.058 |
3.836 ± 0.035 | 7.674 ± 0.049 |
1.883 ± 0.03 | 5.817 ± 0.051 |
4.602 ± 0.053 | 10.115 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.767 ± 0.029 | 3.223 ± 0.036 |
4.056 ± 0.032 | 2.98 ± 0.028 |
6.311 ± 0.072 | 5.494 ± 0.049 |
5.548 ± 0.052 | 7.178 ± 0.047 |
0.846 ± 0.021 | 3.433 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
