
Oenococcus phage phiOE33PA
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
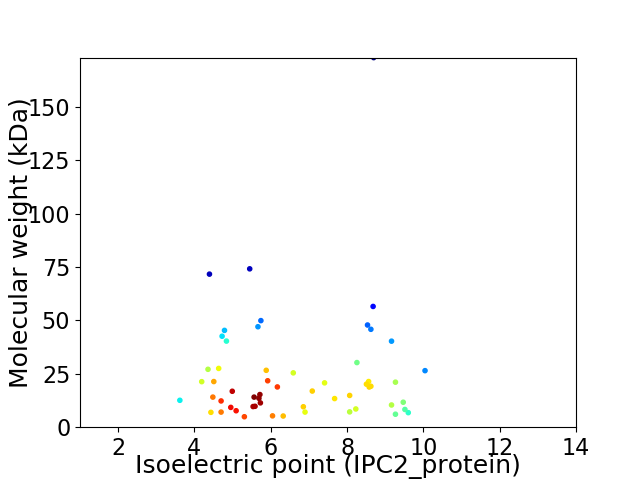
Virtual 2D-PAGE plot for 57 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
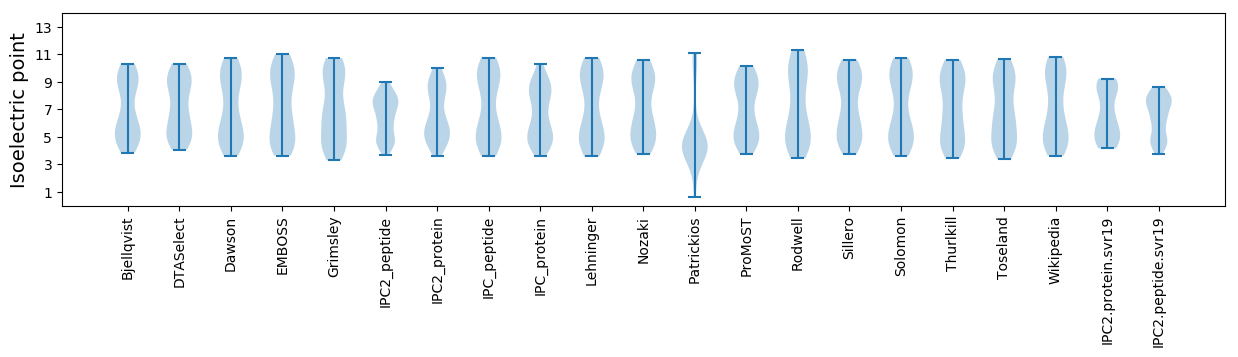
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U9PDS7|A0A2U9PDS7_9CAUD Uncharacterized protein OS=Oenococcus phage phiOE33PA OX=2201414 GN=phiOE33PA_00190 PE=4 SV=1
MM1 pKa = 8.04AITMYY6 pKa = 10.52QADD9 pKa = 4.53RR10 pKa = 11.84NFVSPANDD18 pKa = 3.22AALYY22 pKa = 10.41SAILNNTSGVLANRR36 pKa = 11.84GNNFALTIDD45 pKa = 3.99GLVVSIDD52 pKa = 3.11TGQAVIGGRR61 pKa = 11.84LIEE64 pKa = 4.1ITALEE69 pKa = 4.67SVTVPANSSGSICLVVDD86 pKa = 3.84LTKK89 pKa = 10.91TNTVTGNAGDD99 pKa = 3.77TTYY102 pKa = 11.28SVAVNQVYY110 pKa = 8.69TSAVTGSLTQDD121 pKa = 3.73DD122 pKa = 4.94LNDD125 pKa = 3.54GGFIYY130 pKa = 9.8EE131 pKa = 4.5LPLASFVSTATSVTLTDD148 pKa = 3.23TTGYY152 pKa = 11.07LNDD155 pKa = 4.48TGWLTLPNATGLVIGSGGFTKK176 pKa = 10.67YY177 pKa = 9.58RR178 pKa = 11.84VKK180 pKa = 11.29NNVVYY185 pKa = 10.51IRR187 pKa = 11.84LQALDD192 pKa = 3.51TSKK195 pKa = 7.9TTNANQIGTIPSKK208 pKa = 10.1YY209 pKa = 9.79APSITFMAAGMDD221 pKa = 3.97DD222 pKa = 4.05SSGTPYY228 pKa = 10.75GIEE231 pKa = 3.79LHH233 pKa = 6.0VQTSGLIYY241 pKa = 10.28ANYY244 pKa = 9.79VSPHH248 pKa = 5.81NGGIGGTITYY258 pKa = 9.68PLGG261 pKa = 3.64
MM1 pKa = 8.04AITMYY6 pKa = 10.52QADD9 pKa = 4.53RR10 pKa = 11.84NFVSPANDD18 pKa = 3.22AALYY22 pKa = 10.41SAILNNTSGVLANRR36 pKa = 11.84GNNFALTIDD45 pKa = 3.99GLVVSIDD52 pKa = 3.11TGQAVIGGRR61 pKa = 11.84LIEE64 pKa = 4.1ITALEE69 pKa = 4.67SVTVPANSSGSICLVVDD86 pKa = 3.84LTKK89 pKa = 10.91TNTVTGNAGDD99 pKa = 3.77TTYY102 pKa = 11.28SVAVNQVYY110 pKa = 8.69TSAVTGSLTQDD121 pKa = 3.73DD122 pKa = 4.94LNDD125 pKa = 3.54GGFIYY130 pKa = 9.8EE131 pKa = 4.5LPLASFVSTATSVTLTDD148 pKa = 3.23TTGYY152 pKa = 11.07LNDD155 pKa = 4.48TGWLTLPNATGLVIGSGGFTKK176 pKa = 10.67YY177 pKa = 9.58RR178 pKa = 11.84VKK180 pKa = 11.29NNVVYY185 pKa = 10.51IRR187 pKa = 11.84LQALDD192 pKa = 3.51TSKK195 pKa = 7.9TTNANQIGTIPSKK208 pKa = 10.1YY209 pKa = 9.79APSITFMAAGMDD221 pKa = 3.97DD222 pKa = 4.05SSGTPYY228 pKa = 10.75GIEE231 pKa = 3.79LHH233 pKa = 6.0VQTSGLIYY241 pKa = 10.28ANYY244 pKa = 9.79VSPHH248 pKa = 5.81NGGIGGTITYY258 pKa = 9.68PLGG261 pKa = 3.64
Molecular weight: 27.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U9PF94|A0A2U9PF94_9CAUD HTH-type transcriptional regulator OS=Oenococcus phage phiOE33PA OX=2201414 GN=xre PE=4 SV=1
MM1 pKa = 7.22NVVFLIVIGLIVGTFVILFGGGGAAIYY28 pKa = 10.33LGILTGVVGLNASTAASTSLVTVLPSLILGVWTYY62 pKa = 11.09YY63 pKa = 10.3RR64 pKa = 11.84QGTIKK69 pKa = 9.7TRR71 pKa = 11.84LGNQMLITAIPAVIIGSLISSYY93 pKa = 10.99IPDD96 pKa = 4.33NLYY99 pKa = 10.36KK100 pKa = 10.1WIVGIILILLGINMLFQKK118 pKa = 10.25QKK120 pKa = 9.7SQVDD124 pKa = 3.73PVMSKK129 pKa = 10.4QINRR133 pKa = 11.84KK134 pKa = 9.37DD135 pKa = 3.29RR136 pKa = 11.84FKK138 pKa = 11.37AGIFGIIGGLMVGVAGMSGGAPIIAGLFLIGLSTVNAVATSAYY181 pKa = 10.36VLVFMSAIGTVFHH194 pKa = 7.02IVGSQVDD201 pKa = 3.85WNAGISLMVGALIGAAIGPRR221 pKa = 11.84LLTRR225 pKa = 11.84LTKK228 pKa = 10.65SKK230 pKa = 9.79VGKK233 pKa = 9.78YY234 pKa = 8.8IKK236 pKa = 10.08PVIAFFLVLLGFKK249 pKa = 9.45TLFF252 pKa = 3.78
MM1 pKa = 7.22NVVFLIVIGLIVGTFVILFGGGGAAIYY28 pKa = 10.33LGILTGVVGLNASTAASTSLVTVLPSLILGVWTYY62 pKa = 11.09YY63 pKa = 10.3RR64 pKa = 11.84QGTIKK69 pKa = 9.7TRR71 pKa = 11.84LGNQMLITAIPAVIIGSLISSYY93 pKa = 10.99IPDD96 pKa = 4.33NLYY99 pKa = 10.36KK100 pKa = 10.1WIVGIILILLGINMLFQKK118 pKa = 10.25QKK120 pKa = 9.7SQVDD124 pKa = 3.73PVMSKK129 pKa = 10.4QINRR133 pKa = 11.84KK134 pKa = 9.37DD135 pKa = 3.29RR136 pKa = 11.84FKK138 pKa = 11.37AGIFGIIGGLMVGVAGMSGGAPIIAGLFLIGLSTVNAVATSAYY181 pKa = 10.36VLVFMSAIGTVFHH194 pKa = 7.02IVGSQVDD201 pKa = 3.85WNAGISLMVGALIGAAIGPRR221 pKa = 11.84LLTRR225 pKa = 11.84LTKK228 pKa = 10.65SKK230 pKa = 9.79VGKK233 pKa = 9.78YY234 pKa = 8.8IKK236 pKa = 10.08PVIAFFLVLLGFKK249 pKa = 9.45TLFF252 pKa = 3.78
Molecular weight: 26.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12485 |
44 |
1613 |
219.0 |
24.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.048 ± 0.483 | 0.481 ± 0.109 |
6.552 ± 0.326 | 5.038 ± 0.341 |
4.189 ± 0.316 | 6.063 ± 0.445 |
1.858 ± 0.175 | 7.345 ± 0.372 |
8.474 ± 0.569 | 8.634 ± 0.272 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.243 ± 0.134 | 6.159 ± 0.254 |
2.883 ± 0.245 | 4.405 ± 0.253 |
3.148 ± 0.223 | 7.577 ± 0.454 |
6.696 ± 0.386 | 5.983 ± 0.273 |
1.097 ± 0.131 | 4.125 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
