
Coprococcus eutactus CAG:665
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Coprococcus; environmental samples
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
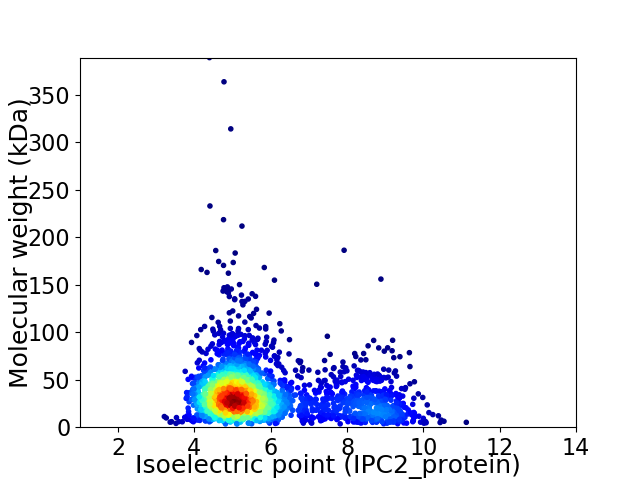
Virtual 2D-PAGE plot for 2415 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5WHH5|R5WHH5_9FIRM Uncharacterized protein OS=Coprococcus eutactus CAG:665 OX=1263071 GN=BN751_01132 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.49KK3 pKa = 10.03KK4 pKa = 10.66GYY6 pKa = 10.5GKK8 pKa = 10.56ALTLLLVTSLVAGTVLTGCGKK29 pKa = 10.62KK30 pKa = 10.15KK31 pKa = 10.25VDD33 pKa = 3.55YY34 pKa = 11.74NMGDD38 pKa = 3.77DD39 pKa = 4.1EE40 pKa = 4.85NGSGGGGKK48 pKa = 9.41LASRR52 pKa = 11.84LDD54 pKa = 3.62VPDD57 pKa = 3.97SYY59 pKa = 11.65EE60 pKa = 4.01GALEE64 pKa = 5.49GIDD67 pKa = 3.57TDD69 pKa = 3.83ATGLTDD75 pKa = 3.76VKK77 pKa = 10.84INASKK82 pKa = 9.04ITVPDD87 pKa = 3.64TDD89 pKa = 3.75KK90 pKa = 11.26MSILYY95 pKa = 9.77YY96 pKa = 10.39NQNQVDD102 pKa = 3.81NEE104 pKa = 4.21YY105 pKa = 10.97KK106 pKa = 10.58KK107 pKa = 10.57RR108 pKa = 11.84VCEE111 pKa = 3.92NFFDD115 pKa = 4.09VSAGVYY121 pKa = 8.34TYY123 pKa = 10.93SWEE126 pKa = 4.09KK127 pKa = 9.96PYY129 pKa = 10.98KK130 pKa = 10.2GDD132 pKa = 4.26LEE134 pKa = 4.72RR135 pKa = 11.84EE136 pKa = 3.83IEE138 pKa = 4.16NYY140 pKa = 10.4EE141 pKa = 4.17EE142 pKa = 4.58LSKK145 pKa = 11.16QSTSDD150 pKa = 3.57DD151 pKa = 3.53EE152 pKa = 5.5KK153 pKa = 11.57SFFDD157 pKa = 5.75DD158 pKa = 4.35YY159 pKa = 11.36ISSLKK164 pKa = 10.2EE165 pKa = 3.75QLKK168 pKa = 8.1TATDD172 pKa = 3.53EE173 pKa = 4.31RR174 pKa = 11.84EE175 pKa = 4.05GAGDD179 pKa = 3.53YY180 pKa = 11.05SADD183 pKa = 3.31AFVGSKK189 pKa = 10.45GEE191 pKa = 3.92NMYY194 pKa = 10.01MISFNSTEE202 pKa = 4.15TGEE205 pKa = 4.28SGGFSIDD212 pKa = 3.98YY213 pKa = 10.13YY214 pKa = 10.6PSDD217 pKa = 3.04QLINYY222 pKa = 7.47RR223 pKa = 11.84PKK225 pKa = 9.79EE226 pKa = 4.12GASSVYY232 pKa = 10.19CYY234 pKa = 10.96SSDD237 pKa = 3.99YY238 pKa = 11.33YY239 pKa = 11.25DD240 pKa = 5.64GEE242 pKa = 4.44DD243 pKa = 3.43TANTATVSQDD253 pKa = 3.36DD254 pKa = 5.15AIQQGLSFLAGCGISDD270 pKa = 4.98IIEE273 pKa = 4.34TGCTDD278 pKa = 6.08LIWEE282 pKa = 4.47YY283 pKa = 11.56SDD285 pKa = 3.24TSYY288 pKa = 9.9NTLASEE294 pKa = 3.99KK295 pKa = 9.13SGYY298 pKa = 10.02VITYY302 pKa = 8.41KK303 pKa = 10.6RR304 pKa = 11.84SVDD307 pKa = 4.03GIAPYY312 pKa = 9.41TPNVYY317 pKa = 10.71NIDD320 pKa = 4.02SLNSSDD326 pKa = 4.88DD327 pKa = 3.22VWYY330 pKa = 8.94DD331 pKa = 3.38TMDD334 pKa = 3.31EE335 pKa = 4.23TFEE338 pKa = 5.03LQIDD342 pKa = 3.9DD343 pKa = 4.11NGIVSAYY350 pKa = 10.2CYY352 pKa = 10.46DD353 pKa = 3.73YY354 pKa = 11.37FKK356 pKa = 11.03ATGDD360 pKa = 3.27KK361 pKa = 10.74KK362 pKa = 11.29EE363 pKa = 4.0NVDD366 pKa = 4.42LISWEE371 pKa = 4.29DD372 pKa = 3.39AVKK375 pKa = 10.6ALPKK379 pKa = 10.42AVNTYY384 pKa = 8.66YY385 pKa = 11.1AEE387 pKa = 4.4NKK389 pKa = 7.57TQYY392 pKa = 11.33SSIEE396 pKa = 4.0FNNVQLAYY404 pKa = 10.81YY405 pKa = 9.49KK406 pKa = 10.54IKK408 pKa = 11.11DD409 pKa = 3.42GDD411 pKa = 3.61KK412 pKa = 10.96YY413 pKa = 11.19EE414 pKa = 4.3YY415 pKa = 10.87LPVWAFAQCEE425 pKa = 4.2KK426 pKa = 10.49TGDD429 pKa = 3.81GDD431 pKa = 4.13SSQSEE436 pKa = 4.45DD437 pKa = 3.71GLDD440 pKa = 3.42VSNPSQLIMLNAEE453 pKa = 3.97TGEE456 pKa = 4.44LIDD459 pKa = 5.36LKK461 pKa = 11.12SVLNTQSFSYY471 pKa = 10.46TDD473 pKa = 3.44STVVGGDD480 pKa = 3.79DD481 pKa = 5.46DD482 pKa = 5.73NVTLDD487 pKa = 5.61DD488 pKa = 4.8SDD490 pKa = 5.1LDD492 pKa = 3.85EE493 pKa = 6.55DD494 pKa = 4.44GASIADD500 pKa = 3.97DD501 pKa = 3.78SAVSDD506 pKa = 4.54DD507 pKa = 5.01SSDD510 pKa = 3.68SSDD513 pKa = 4.93DD514 pKa = 3.53GSIDD518 pKa = 4.4INDD521 pKa = 4.91LDD523 pKa = 4.57LQVDD527 pKa = 3.93DD528 pKa = 4.06TTDD531 pKa = 3.2AVEE534 pKa = 4.02EE535 pKa = 4.23
MM1 pKa = 7.43KK2 pKa = 10.49KK3 pKa = 10.03KK4 pKa = 10.66GYY6 pKa = 10.5GKK8 pKa = 10.56ALTLLLVTSLVAGTVLTGCGKK29 pKa = 10.62KK30 pKa = 10.15KK31 pKa = 10.25VDD33 pKa = 3.55YY34 pKa = 11.74NMGDD38 pKa = 3.77DD39 pKa = 4.1EE40 pKa = 4.85NGSGGGGKK48 pKa = 9.41LASRR52 pKa = 11.84LDD54 pKa = 3.62VPDD57 pKa = 3.97SYY59 pKa = 11.65EE60 pKa = 4.01GALEE64 pKa = 5.49GIDD67 pKa = 3.57TDD69 pKa = 3.83ATGLTDD75 pKa = 3.76VKK77 pKa = 10.84INASKK82 pKa = 9.04ITVPDD87 pKa = 3.64TDD89 pKa = 3.75KK90 pKa = 11.26MSILYY95 pKa = 9.77YY96 pKa = 10.39NQNQVDD102 pKa = 3.81NEE104 pKa = 4.21YY105 pKa = 10.97KK106 pKa = 10.58KK107 pKa = 10.57RR108 pKa = 11.84VCEE111 pKa = 3.92NFFDD115 pKa = 4.09VSAGVYY121 pKa = 8.34TYY123 pKa = 10.93SWEE126 pKa = 4.09KK127 pKa = 9.96PYY129 pKa = 10.98KK130 pKa = 10.2GDD132 pKa = 4.26LEE134 pKa = 4.72RR135 pKa = 11.84EE136 pKa = 3.83IEE138 pKa = 4.16NYY140 pKa = 10.4EE141 pKa = 4.17EE142 pKa = 4.58LSKK145 pKa = 11.16QSTSDD150 pKa = 3.57DD151 pKa = 3.53EE152 pKa = 5.5KK153 pKa = 11.57SFFDD157 pKa = 5.75DD158 pKa = 4.35YY159 pKa = 11.36ISSLKK164 pKa = 10.2EE165 pKa = 3.75QLKK168 pKa = 8.1TATDD172 pKa = 3.53EE173 pKa = 4.31RR174 pKa = 11.84EE175 pKa = 4.05GAGDD179 pKa = 3.53YY180 pKa = 11.05SADD183 pKa = 3.31AFVGSKK189 pKa = 10.45GEE191 pKa = 3.92NMYY194 pKa = 10.01MISFNSTEE202 pKa = 4.15TGEE205 pKa = 4.28SGGFSIDD212 pKa = 3.98YY213 pKa = 10.13YY214 pKa = 10.6PSDD217 pKa = 3.04QLINYY222 pKa = 7.47RR223 pKa = 11.84PKK225 pKa = 9.79EE226 pKa = 4.12GASSVYY232 pKa = 10.19CYY234 pKa = 10.96SSDD237 pKa = 3.99YY238 pKa = 11.33YY239 pKa = 11.25DD240 pKa = 5.64GEE242 pKa = 4.44DD243 pKa = 3.43TANTATVSQDD253 pKa = 3.36DD254 pKa = 5.15AIQQGLSFLAGCGISDD270 pKa = 4.98IIEE273 pKa = 4.34TGCTDD278 pKa = 6.08LIWEE282 pKa = 4.47YY283 pKa = 11.56SDD285 pKa = 3.24TSYY288 pKa = 9.9NTLASEE294 pKa = 3.99KK295 pKa = 9.13SGYY298 pKa = 10.02VITYY302 pKa = 8.41KK303 pKa = 10.6RR304 pKa = 11.84SVDD307 pKa = 4.03GIAPYY312 pKa = 9.41TPNVYY317 pKa = 10.71NIDD320 pKa = 4.02SLNSSDD326 pKa = 4.88DD327 pKa = 3.22VWYY330 pKa = 8.94DD331 pKa = 3.38TMDD334 pKa = 3.31EE335 pKa = 4.23TFEE338 pKa = 5.03LQIDD342 pKa = 3.9DD343 pKa = 4.11NGIVSAYY350 pKa = 10.2CYY352 pKa = 10.46DD353 pKa = 3.73YY354 pKa = 11.37FKK356 pKa = 11.03ATGDD360 pKa = 3.27KK361 pKa = 10.74KK362 pKa = 11.29EE363 pKa = 4.0NVDD366 pKa = 4.42LISWEE371 pKa = 4.29DD372 pKa = 3.39AVKK375 pKa = 10.6ALPKK379 pKa = 10.42AVNTYY384 pKa = 8.66YY385 pKa = 11.1AEE387 pKa = 4.4NKK389 pKa = 7.57TQYY392 pKa = 11.33SSIEE396 pKa = 4.0FNNVQLAYY404 pKa = 10.81YY405 pKa = 9.49KK406 pKa = 10.54IKK408 pKa = 11.11DD409 pKa = 3.42GDD411 pKa = 3.61KK412 pKa = 10.96YY413 pKa = 11.19EE414 pKa = 4.3YY415 pKa = 10.87LPVWAFAQCEE425 pKa = 4.2KK426 pKa = 10.49TGDD429 pKa = 3.81GDD431 pKa = 4.13SSQSEE436 pKa = 4.45DD437 pKa = 3.71GLDD440 pKa = 3.42VSNPSQLIMLNAEE453 pKa = 3.97TGEE456 pKa = 4.44LIDD459 pKa = 5.36LKK461 pKa = 11.12SVLNTQSFSYY471 pKa = 10.46TDD473 pKa = 3.44STVVGGDD480 pKa = 3.79DD481 pKa = 5.46DD482 pKa = 5.73NVTLDD487 pKa = 5.61DD488 pKa = 4.8SDD490 pKa = 5.1LDD492 pKa = 3.85EE493 pKa = 6.55DD494 pKa = 4.44GASIADD500 pKa = 3.97DD501 pKa = 3.78SAVSDD506 pKa = 4.54DD507 pKa = 5.01SSDD510 pKa = 3.68SSDD513 pKa = 4.93DD514 pKa = 3.53GSIDD518 pKa = 4.4INDD521 pKa = 4.91LDD523 pKa = 4.57LQVDD527 pKa = 3.93DD528 pKa = 4.06TTDD531 pKa = 3.2AVEE534 pKa = 4.02EE535 pKa = 4.23
Molecular weight: 58.81 kDa
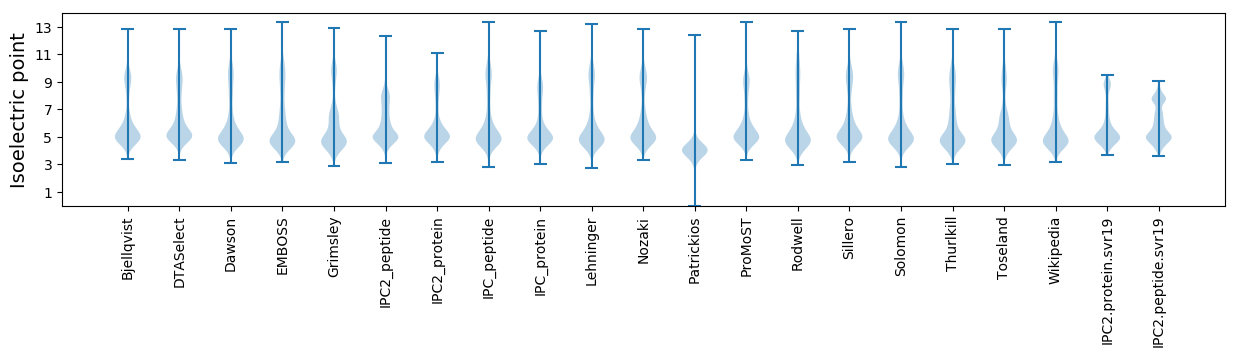
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5VRR7|R5VRR7_9FIRM Putative DNA metabolism protein OS=Coprococcus eutactus CAG:665 OX=1263071 GN=BN751_02319 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
797336 |
30 |
3567 |
330.2 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.279 ± 0.045 | 1.509 ± 0.022 |
6.923 ± 0.049 | 6.81 ± 0.051 |
3.843 ± 0.039 | 7.103 ± 0.044 |
1.58 ± 0.02 | 7.618 ± 0.052 |
6.976 ± 0.047 | 7.922 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.212 ± 0.032 | 4.724 ± 0.038 |
2.909 ± 0.028 | 2.717 ± 0.028 |
4.281 ± 0.041 | 6.243 ± 0.044 |
5.542 ± 0.051 | 7.416 ± 0.041 |
0.839 ± 0.017 | 4.543 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
