
Wuhan House Fly Virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alphadrosrhavirus; Hubei alphadrosrhavirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
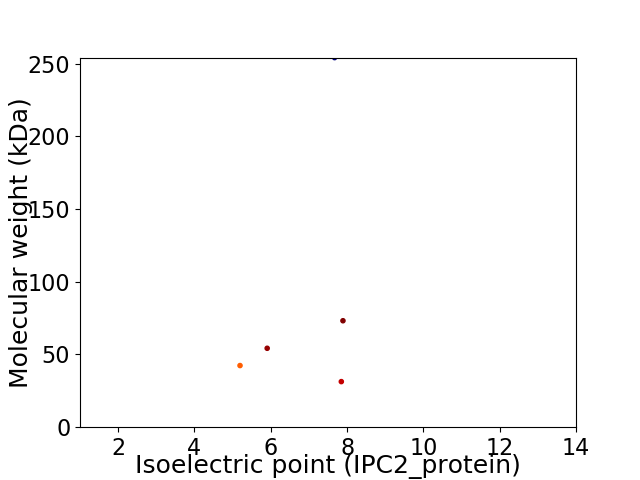
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KRN2|A0A0B5KRN2_9RHAB ORF3 OS=Wuhan House Fly Virus 2 OX=1608105 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.45SMCTFQPCVMGAAAHH16 pKa = 6.26VYY18 pKa = 10.03NADD21 pKa = 3.0KK22 pKa = 10.63CAPAEE27 pKa = 4.27SPFGSEE33 pKa = 4.05DD34 pKa = 3.35DD35 pKa = 3.63QAIAEE40 pKa = 4.64GPDD43 pKa = 3.42TLEE46 pKa = 4.4LDD48 pKa = 3.78SVTSYY53 pKa = 11.41NSLMRR58 pKa = 11.84DD59 pKa = 4.34FYY61 pKa = 11.39AGEE64 pKa = 4.08PTTLHH69 pKa = 6.41SEE71 pKa = 4.2LDD73 pKa = 3.51ADD75 pKa = 4.69TVCNMPTISEE85 pKa = 4.48EE86 pKa = 3.91IFGNASSPKK95 pKa = 9.22EE96 pKa = 3.93NVSSSRR102 pKa = 11.84EE103 pKa = 3.6IIAFLDD109 pKa = 3.19SWTSFTEE116 pKa = 3.74INQLKK121 pKa = 9.8EE122 pKa = 3.88SPHH125 pKa = 6.3YY126 pKa = 10.37FFEE129 pKa = 4.75KK130 pKa = 9.26AQIMIKK136 pKa = 10.64APDD139 pKa = 3.66TLKK142 pKa = 10.94EE143 pKa = 3.89IMSGFRR149 pKa = 11.84VLAQVNTALLTEE161 pKa = 4.35FLLRR165 pKa = 11.84IVGSSNPDD173 pKa = 3.83DD174 pKa = 3.7IKK176 pKa = 10.92MFSALARR183 pKa = 11.84GAMYY187 pKa = 10.32ISSGFQKK194 pKa = 10.9LEE196 pKa = 3.91CTKK199 pKa = 10.55SYY201 pKa = 10.92EE202 pKa = 4.16ILSHH206 pKa = 6.85NMEE209 pKa = 5.15LLTHH213 pKa = 6.48HH214 pKa = 6.68VSAVKK219 pKa = 10.82DD220 pKa = 3.77NVTSQSDD227 pKa = 3.58MVEE230 pKa = 4.12SLRR233 pKa = 11.84EE234 pKa = 4.01SVNKK238 pKa = 9.74VVSVTSKK245 pKa = 11.31LEE247 pKa = 4.17AEE249 pKa = 4.62AKK251 pKa = 10.76LLDD254 pKa = 3.5IKK256 pKa = 10.64AAKK259 pKa = 9.6NSPYY263 pKa = 9.75HH264 pKa = 6.6ASSLPSSSSVGTPIQITGYY283 pKa = 10.01KK284 pKa = 8.92CDD286 pKa = 3.55PHH288 pKa = 8.63IIVCNKK294 pKa = 8.75EE295 pKa = 3.8GKK297 pKa = 10.04VDD299 pKa = 3.83VEE301 pKa = 4.33ATLKK305 pKa = 10.86ANKK308 pKa = 9.54DD309 pKa = 3.19LSSIHH314 pKa = 6.94KK315 pKa = 8.68KK316 pKa = 8.31TLDD319 pKa = 3.26VLALFKK325 pKa = 10.43PAALQFLMAVDD336 pKa = 4.26WSTLNSLMPVDD347 pKa = 4.69PEE349 pKa = 4.78LSVKK353 pKa = 10.03QAGEE357 pKa = 3.53MLYY360 pKa = 10.17TKK362 pKa = 10.41FRR364 pKa = 11.84SLYY367 pKa = 7.9GNRR370 pKa = 11.84MPRR373 pKa = 11.84QPEE376 pKa = 4.2TLTLRR381 pKa = 11.84QGKK384 pKa = 8.35NN385 pKa = 2.84
MM1 pKa = 7.45SMCTFQPCVMGAAAHH16 pKa = 6.26VYY18 pKa = 10.03NADD21 pKa = 3.0KK22 pKa = 10.63CAPAEE27 pKa = 4.27SPFGSEE33 pKa = 4.05DD34 pKa = 3.35DD35 pKa = 3.63QAIAEE40 pKa = 4.64GPDD43 pKa = 3.42TLEE46 pKa = 4.4LDD48 pKa = 3.78SVTSYY53 pKa = 11.41NSLMRR58 pKa = 11.84DD59 pKa = 4.34FYY61 pKa = 11.39AGEE64 pKa = 4.08PTTLHH69 pKa = 6.41SEE71 pKa = 4.2LDD73 pKa = 3.51ADD75 pKa = 4.69TVCNMPTISEE85 pKa = 4.48EE86 pKa = 3.91IFGNASSPKK95 pKa = 9.22EE96 pKa = 3.93NVSSSRR102 pKa = 11.84EE103 pKa = 3.6IIAFLDD109 pKa = 3.19SWTSFTEE116 pKa = 3.74INQLKK121 pKa = 9.8EE122 pKa = 3.88SPHH125 pKa = 6.3YY126 pKa = 10.37FFEE129 pKa = 4.75KK130 pKa = 9.26AQIMIKK136 pKa = 10.64APDD139 pKa = 3.66TLKK142 pKa = 10.94EE143 pKa = 3.89IMSGFRR149 pKa = 11.84VLAQVNTALLTEE161 pKa = 4.35FLLRR165 pKa = 11.84IVGSSNPDD173 pKa = 3.83DD174 pKa = 3.7IKK176 pKa = 10.92MFSALARR183 pKa = 11.84GAMYY187 pKa = 10.32ISSGFQKK194 pKa = 10.9LEE196 pKa = 3.91CTKK199 pKa = 10.55SYY201 pKa = 10.92EE202 pKa = 4.16ILSHH206 pKa = 6.85NMEE209 pKa = 5.15LLTHH213 pKa = 6.48HH214 pKa = 6.68VSAVKK219 pKa = 10.82DD220 pKa = 3.77NVTSQSDD227 pKa = 3.58MVEE230 pKa = 4.12SLRR233 pKa = 11.84EE234 pKa = 4.01SVNKK238 pKa = 9.74VVSVTSKK245 pKa = 11.31LEE247 pKa = 4.17AEE249 pKa = 4.62AKK251 pKa = 10.76LLDD254 pKa = 3.5IKK256 pKa = 10.64AAKK259 pKa = 9.6NSPYY263 pKa = 9.75HH264 pKa = 6.6ASSLPSSSSVGTPIQITGYY283 pKa = 10.01KK284 pKa = 8.92CDD286 pKa = 3.55PHH288 pKa = 8.63IIVCNKK294 pKa = 8.75EE295 pKa = 3.8GKK297 pKa = 10.04VDD299 pKa = 3.83VEE301 pKa = 4.33ATLKK305 pKa = 10.86ANKK308 pKa = 9.54DD309 pKa = 3.19LSSIHH314 pKa = 6.94KK315 pKa = 8.68KK316 pKa = 8.31TLDD319 pKa = 3.26VLALFKK325 pKa = 10.43PAALQFLMAVDD336 pKa = 4.26WSTLNSLMPVDD347 pKa = 4.69PEE349 pKa = 4.78LSVKK353 pKa = 10.03QAGEE357 pKa = 3.53MLYY360 pKa = 10.17TKK362 pKa = 10.41FRR364 pKa = 11.84SLYY367 pKa = 7.9GNRR370 pKa = 11.84MPRR373 pKa = 11.84QPEE376 pKa = 4.2TLTLRR381 pKa = 11.84QGKK384 pKa = 8.35NN385 pKa = 2.84
Molecular weight: 42.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KTL0|A0A0B5KTL0_9RHAB Glycoprotein OS=Wuhan House Fly Virus 2 OX=1608105 GN=G PE=4 SV=1
MM1 pKa = 7.73SDD3 pKa = 4.34NIPSHH8 pKa = 5.26SAGRR12 pKa = 11.84IDD14 pKa = 3.83KK15 pKa = 10.3VEE17 pKa = 4.66HH18 pKa = 6.68IISDD22 pKa = 3.61FTEE25 pKa = 4.4QISLILQGTIEE36 pKa = 4.32LSGPEE41 pKa = 4.0CDD43 pKa = 3.77YY44 pKa = 11.33SIFMWRR50 pKa = 11.84VCCAIAYY57 pKa = 8.11DD58 pKa = 3.64VTYY61 pKa = 10.39KK62 pKa = 10.86KK63 pKa = 10.57KK64 pKa = 8.95VTRR67 pKa = 11.84NTQYY71 pKa = 11.38LIASLIYY78 pKa = 10.24QSLRR82 pKa = 11.84CHH84 pKa = 5.88SQLFSSLQPHH94 pKa = 6.57YY95 pKa = 11.04GPLLEE100 pKa = 4.7LVCPIKK106 pKa = 10.91VRR108 pKa = 11.84GYY110 pKa = 8.14TEE112 pKa = 3.97YY113 pKa = 10.35PIEE116 pKa = 4.26TRR118 pKa = 11.84SFPQLNIIRR127 pKa = 11.84EE128 pKa = 3.88RR129 pKa = 11.84SFRR132 pKa = 11.84YY133 pKa = 9.33QVTDD137 pKa = 3.16VKK139 pKa = 10.3QLRR142 pKa = 11.84IRR144 pKa = 11.84SRR146 pKa = 11.84IQLQACQFSDD156 pKa = 3.21NFRR159 pKa = 11.84AEE161 pKa = 4.07ARR163 pKa = 11.84LKK165 pKa = 10.8GYY167 pKa = 10.52VPLLPPSTDD176 pKa = 2.4EE177 pKa = 4.15WIVYY181 pKa = 9.61FSEE184 pKa = 4.55LEE186 pKa = 3.94NAIDD190 pKa = 3.36STGRR194 pKa = 11.84YY195 pKa = 8.88DD196 pKa = 3.97QIDD199 pKa = 3.93VSAHH203 pKa = 4.84IAKK206 pKa = 10.25GIEE209 pKa = 3.89ALTPSKK215 pKa = 10.58PNEE218 pKa = 4.01SVSFVKK224 pKa = 10.5KK225 pKa = 10.75LKK227 pKa = 10.52QFMSKK232 pKa = 9.7SSKK235 pKa = 9.71NAVVEE240 pKa = 4.2ARR242 pKa = 11.84ALAPSHH248 pKa = 7.04PSDD251 pKa = 3.73CLEE254 pKa = 4.78IISSPKK260 pKa = 10.02VVNQQVILSRR270 pKa = 11.84QPKK273 pKa = 9.56NNN275 pKa = 3.31
MM1 pKa = 7.73SDD3 pKa = 4.34NIPSHH8 pKa = 5.26SAGRR12 pKa = 11.84IDD14 pKa = 3.83KK15 pKa = 10.3VEE17 pKa = 4.66HH18 pKa = 6.68IISDD22 pKa = 3.61FTEE25 pKa = 4.4QISLILQGTIEE36 pKa = 4.32LSGPEE41 pKa = 4.0CDD43 pKa = 3.77YY44 pKa = 11.33SIFMWRR50 pKa = 11.84VCCAIAYY57 pKa = 8.11DD58 pKa = 3.64VTYY61 pKa = 10.39KK62 pKa = 10.86KK63 pKa = 10.57KK64 pKa = 8.95VTRR67 pKa = 11.84NTQYY71 pKa = 11.38LIASLIYY78 pKa = 10.24QSLRR82 pKa = 11.84CHH84 pKa = 5.88SQLFSSLQPHH94 pKa = 6.57YY95 pKa = 11.04GPLLEE100 pKa = 4.7LVCPIKK106 pKa = 10.91VRR108 pKa = 11.84GYY110 pKa = 8.14TEE112 pKa = 3.97YY113 pKa = 10.35PIEE116 pKa = 4.26TRR118 pKa = 11.84SFPQLNIIRR127 pKa = 11.84EE128 pKa = 3.88RR129 pKa = 11.84SFRR132 pKa = 11.84YY133 pKa = 9.33QVTDD137 pKa = 3.16VKK139 pKa = 10.3QLRR142 pKa = 11.84IRR144 pKa = 11.84SRR146 pKa = 11.84IQLQACQFSDD156 pKa = 3.21NFRR159 pKa = 11.84AEE161 pKa = 4.07ARR163 pKa = 11.84LKK165 pKa = 10.8GYY167 pKa = 10.52VPLLPPSTDD176 pKa = 2.4EE177 pKa = 4.15WIVYY181 pKa = 9.61FSEE184 pKa = 4.55LEE186 pKa = 3.94NAIDD190 pKa = 3.36STGRR194 pKa = 11.84YY195 pKa = 8.88DD196 pKa = 3.97QIDD199 pKa = 3.93VSAHH203 pKa = 4.84IAKK206 pKa = 10.25GIEE209 pKa = 3.89ALTPSKK215 pKa = 10.58PNEE218 pKa = 4.01SVSFVKK224 pKa = 10.5KK225 pKa = 10.75LKK227 pKa = 10.52QFMSKK232 pKa = 9.7SSKK235 pKa = 9.71NAVVEE240 pKa = 4.2ARR242 pKa = 11.84ALAPSHH248 pKa = 7.04PSDD251 pKa = 3.73CLEE254 pKa = 4.78IISSPKK260 pKa = 10.02VVNQQVILSRR270 pKa = 11.84QPKK273 pKa = 9.56NNN275 pKa = 3.31
Molecular weight: 31.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4031 |
275 |
2233 |
806.2 |
91.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.483 ± 0.677 | 1.96 ± 0.133 |
4.986 ± 0.397 | 5.408 ± 0.252 |
5.11 ± 0.544 | 4.838 ± 0.328 |
2.506 ± 0.082 | 7.12 ± 0.336 |
6.773 ± 0.145 | 10.146 ± 0.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.803 ± 0.233 | 4.589 ± 0.251 |
5.036 ± 0.185 | 3.597 ± 0.407 |
4.118 ± 0.302 | 9.799 ± 0.546 |
5.334 ± 0.247 | 5.83 ± 0.149 |
1.439 ± 0.282 | 3.126 ± 0.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
