
Shimia sp. SK013
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Shimia; unclassified Shimia
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
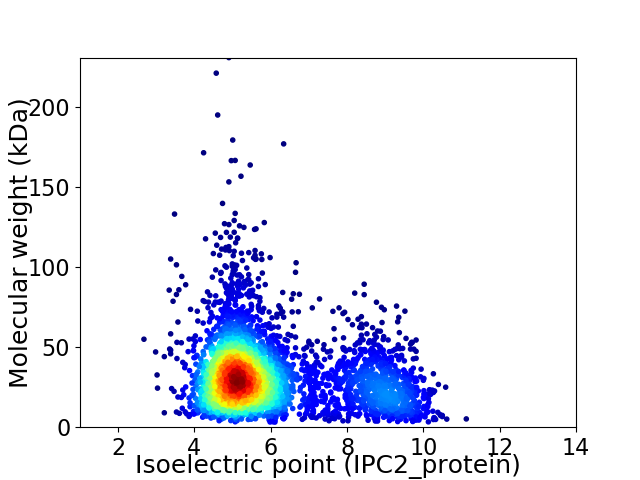
Virtual 2D-PAGE plot for 3979 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
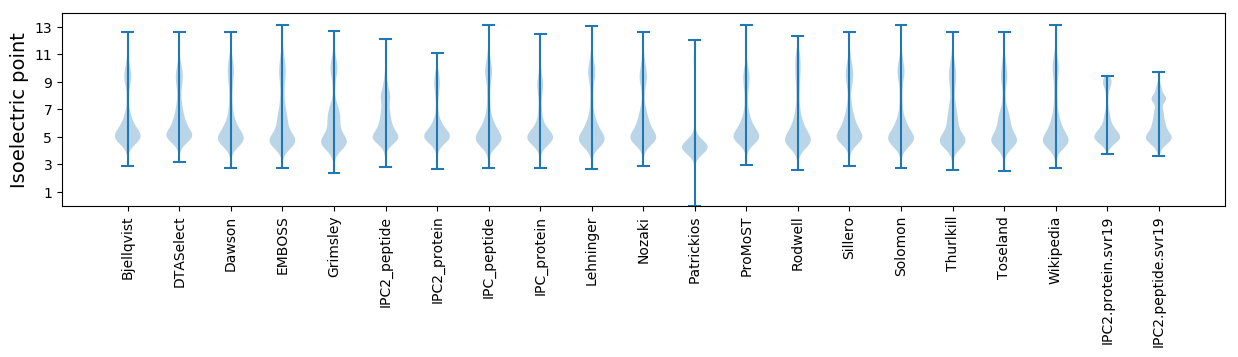
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M9EK91|A0A0M9EK91_9RHOB Murein hydrolase activator NlpD OS=Shimia sp. SK013 OX=1389006 GN=nlpD PE=4 SV=1
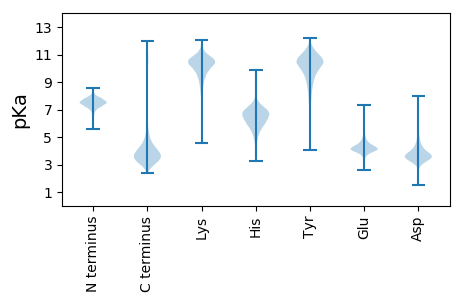
MM1 pKa = 7.23PTYY4 pKa = 9.89FQHH7 pKa = 5.68EE8 pKa = 4.78QYY10 pKa = 11.45GLAHH14 pKa = 7.17LFSEE18 pKa = 4.88HH19 pKa = 7.0FADD22 pKa = 3.67RR23 pKa = 11.84WNTAFVTEE31 pKa = 4.27TMNGEE36 pKa = 3.97VRR38 pKa = 11.84IGFSPSSPIVYY49 pKa = 10.11GVMQLNGDD57 pKa = 4.15DD58 pKa = 3.72LVVINGEE65 pKa = 4.53LISGTITSIDD75 pKa = 3.9FYY77 pKa = 11.38YY78 pKa = 10.13PGYY81 pKa = 9.7PSGGSFGVGFLDD93 pKa = 4.78GQLTGLNWGAQDD105 pKa = 4.13FQSQLAAFAAGDD117 pKa = 3.62YY118 pKa = 11.06SGFNQAILDD127 pKa = 3.94TVTTVICVDD136 pKa = 3.44TRR138 pKa = 11.84PTVHH142 pKa = 6.78SGDD145 pKa = 3.71YY146 pKa = 11.09ANANSFVLGNNDD158 pKa = 2.9GTFSFEE164 pKa = 3.76HH165 pKa = 6.3SALGTSVPITINGGHH180 pKa = 6.85GYY182 pKa = 10.49DD183 pKa = 3.1IFSVSWYY190 pKa = 9.94VPPPTPLSPAPTFTIDD206 pKa = 4.02LEE208 pKa = 5.26AGTFTDD214 pKa = 5.74LYY216 pKa = 9.32GTTHH220 pKa = 7.48RR221 pKa = 11.84ILNFEE226 pKa = 4.12EE227 pKa = 4.86VIGSSANNHH236 pKa = 3.89ITGYY240 pKa = 10.28EE241 pKa = 4.04NVSNALTGGVGDD253 pKa = 4.25DD254 pKa = 3.8TLLGGNQSDD263 pKa = 4.06TLVGGSGNDD272 pKa = 3.5LLVAGDD278 pKa = 4.01GDD280 pKa = 4.4DD281 pKa = 5.09VIMGSDD287 pKa = 3.63NFSDD291 pKa = 5.71DD292 pKa = 4.7DD293 pKa = 3.85DD294 pKa = 4.93TINGGDD300 pKa = 3.95GIDD303 pKa = 3.76TYY305 pKa = 10.22RR306 pKa = 11.84TSYY309 pKa = 11.15SSDD312 pKa = 3.02HH313 pKa = 6.76FVDD316 pKa = 4.07LTNTYY321 pKa = 7.75STPHH325 pKa = 6.09GNTRR329 pKa = 11.84TFIDD333 pKa = 3.47IEE335 pKa = 4.27NVITRR340 pKa = 11.84SGDD343 pKa = 3.17DD344 pKa = 3.6TIIGNGVDD352 pKa = 3.69NRR354 pKa = 11.84IEE356 pKa = 3.93SWAGEE361 pKa = 4.06DD362 pKa = 4.92LILAGGGNDD371 pKa = 3.53TVIGGSGNDD380 pKa = 3.21ILVGGTGEE388 pKa = 4.14NEE390 pKa = 3.9LTGGRR395 pKa = 11.84DD396 pKa = 2.88EE397 pKa = 4.57DD398 pKa = 3.61QFVFVTLNASDD409 pKa = 4.62TITDD413 pKa = 3.97FTMSEE418 pKa = 4.06DD419 pKa = 3.69TLDD422 pKa = 4.08FSSLAADD429 pKa = 3.33SGIFSVTMNGLEE441 pKa = 4.4TIRR444 pKa = 11.84INCFDD449 pKa = 4.0DD450 pKa = 3.63TASGILALSIVQSGDD465 pKa = 2.91AVEE468 pKa = 4.96IYY470 pKa = 9.1VTSGGIAASGSTPLATLLDD489 pKa = 3.97VNLADD494 pKa = 5.72LSWDD498 pKa = 3.52DD499 pKa = 5.39FIFF502 pKa = 4.5
MM1 pKa = 7.23PTYY4 pKa = 9.89FQHH7 pKa = 5.68EE8 pKa = 4.78QYY10 pKa = 11.45GLAHH14 pKa = 7.17LFSEE18 pKa = 4.88HH19 pKa = 7.0FADD22 pKa = 3.67RR23 pKa = 11.84WNTAFVTEE31 pKa = 4.27TMNGEE36 pKa = 3.97VRR38 pKa = 11.84IGFSPSSPIVYY49 pKa = 10.11GVMQLNGDD57 pKa = 4.15DD58 pKa = 3.72LVVINGEE65 pKa = 4.53LISGTITSIDD75 pKa = 3.9FYY77 pKa = 11.38YY78 pKa = 10.13PGYY81 pKa = 9.7PSGGSFGVGFLDD93 pKa = 4.78GQLTGLNWGAQDD105 pKa = 4.13FQSQLAAFAAGDD117 pKa = 3.62YY118 pKa = 11.06SGFNQAILDD127 pKa = 3.94TVTTVICVDD136 pKa = 3.44TRR138 pKa = 11.84PTVHH142 pKa = 6.78SGDD145 pKa = 3.71YY146 pKa = 11.09ANANSFVLGNNDD158 pKa = 2.9GTFSFEE164 pKa = 3.76HH165 pKa = 6.3SALGTSVPITINGGHH180 pKa = 6.85GYY182 pKa = 10.49DD183 pKa = 3.1IFSVSWYY190 pKa = 9.94VPPPTPLSPAPTFTIDD206 pKa = 4.02LEE208 pKa = 5.26AGTFTDD214 pKa = 5.74LYY216 pKa = 9.32GTTHH220 pKa = 7.48RR221 pKa = 11.84ILNFEE226 pKa = 4.12EE227 pKa = 4.86VIGSSANNHH236 pKa = 3.89ITGYY240 pKa = 10.28EE241 pKa = 4.04NVSNALTGGVGDD253 pKa = 4.25DD254 pKa = 3.8TLLGGNQSDD263 pKa = 4.06TLVGGSGNDD272 pKa = 3.5LLVAGDD278 pKa = 4.01GDD280 pKa = 4.4DD281 pKa = 5.09VIMGSDD287 pKa = 3.63NFSDD291 pKa = 5.71DD292 pKa = 4.7DD293 pKa = 3.85DD294 pKa = 4.93TINGGDD300 pKa = 3.95GIDD303 pKa = 3.76TYY305 pKa = 10.22RR306 pKa = 11.84TSYY309 pKa = 11.15SSDD312 pKa = 3.02HH313 pKa = 6.76FVDD316 pKa = 4.07LTNTYY321 pKa = 7.75STPHH325 pKa = 6.09GNTRR329 pKa = 11.84TFIDD333 pKa = 3.47IEE335 pKa = 4.27NVITRR340 pKa = 11.84SGDD343 pKa = 3.17DD344 pKa = 3.6TIIGNGVDD352 pKa = 3.69NRR354 pKa = 11.84IEE356 pKa = 3.93SWAGEE361 pKa = 4.06DD362 pKa = 4.92LILAGGGNDD371 pKa = 3.53TVIGGSGNDD380 pKa = 3.21ILVGGTGEE388 pKa = 4.14NEE390 pKa = 3.9LTGGRR395 pKa = 11.84DD396 pKa = 2.88EE397 pKa = 4.57DD398 pKa = 3.61QFVFVTLNASDD409 pKa = 4.62TITDD413 pKa = 3.97FTMSEE418 pKa = 4.06DD419 pKa = 3.69TLDD422 pKa = 4.08FSSLAADD429 pKa = 3.33SGIFSVTMNGLEE441 pKa = 4.4TIRR444 pKa = 11.84INCFDD449 pKa = 4.0DD450 pKa = 3.63TASGILALSIVQSGDD465 pKa = 2.91AVEE468 pKa = 4.96IYY470 pKa = 9.1VTSGGIAASGSTPLATLLDD489 pKa = 3.97VNLADD494 pKa = 5.72LSWDD498 pKa = 3.52DD499 pKa = 5.39FIFF502 pKa = 4.5
Molecular weight: 52.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M9EFS2|A0A0M9EFS2_9RHOB Pseudouridine synthase OS=Shimia sp. SK013 OX=1389006 GN=rluC PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1221488 |
29 |
2145 |
307.0 |
33.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.701 ± 0.041 | 0.907 ± 0.013 |
6.265 ± 0.036 | 5.818 ± 0.034 |
3.957 ± 0.023 | 8.536 ± 0.042 |
2.098 ± 0.021 | 5.188 ± 0.029 |
3.631 ± 0.03 | 9.767 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.964 ± 0.017 | 2.907 ± 0.02 |
4.749 ± 0.025 | 3.261 ± 0.019 |
6.015 ± 0.033 | 5.428 ± 0.025 |
5.626 ± 0.028 | 7.557 ± 0.035 |
1.39 ± 0.016 | 2.234 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
