
Gata virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
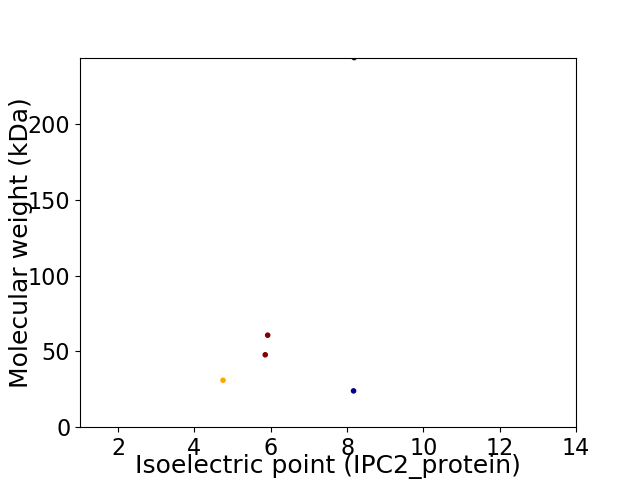
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z2CF63|A0A2Z2CF63_9RHAB Glycoprotein OS=Gata virus OX=1911435 PE=3 SV=1
MM1 pKa = 7.79KK2 pKa = 9.17KK3 pKa = 7.62TQQHH7 pKa = 5.05SLRR10 pKa = 11.84PIMDD14 pKa = 3.79EE15 pKa = 3.9LNEE18 pKa = 4.09KK19 pKa = 10.13FKK21 pKa = 11.35QFLDD25 pKa = 3.65DD26 pKa = 3.95GARR29 pKa = 11.84QNILSDD35 pKa = 3.37EE36 pKa = 4.63FKK38 pKa = 10.85RR39 pKa = 11.84DD40 pKa = 3.08VNAILGEE47 pKa = 4.18MEE49 pKa = 4.55DD50 pKa = 4.19EE51 pKa = 4.39EE52 pKa = 5.05MPLAPNDD59 pKa = 3.8HH60 pKa = 6.38SPVRR64 pKa = 11.84QSTSLYY70 pKa = 10.93SPDD73 pKa = 4.62HH74 pKa = 7.08DD75 pKa = 5.13LDD77 pKa = 3.98TYY79 pKa = 10.9TDD81 pKa = 4.42PGCQSPPGSVCDD93 pKa = 4.13PLDD96 pKa = 5.06DD97 pKa = 4.23EE98 pKa = 5.11ACSILGEE105 pKa = 4.12LEE107 pKa = 4.47RR108 pKa = 11.84IVTLSDD114 pKa = 4.7PGNQWEE120 pKa = 4.62GASHH124 pKa = 6.92IVWTSPGVYY133 pKa = 8.46STGRR137 pKa = 11.84EE138 pKa = 3.82FQQAKK143 pKa = 8.5TLLKK147 pKa = 10.57SVLRR151 pKa = 11.84SMNRR155 pKa = 11.84DD156 pKa = 2.86SGLNYY161 pKa = 9.8VLNVGKK167 pKa = 9.58PDD169 pKa = 3.41TLFITRR175 pKa = 11.84PDD177 pKa = 3.48SLTVARR183 pKa = 11.84NVRR186 pKa = 11.84PKK188 pKa = 10.16IPEE191 pKa = 3.83NLFVDD196 pKa = 3.43NDD198 pKa = 2.84KK199 pKa = 11.22WEE201 pKa = 4.75FIKK204 pKa = 11.03AQFASGIEE212 pKa = 4.09LKK214 pKa = 10.96DD215 pKa = 3.09VDD217 pKa = 4.6EE218 pKa = 4.9PGKK221 pKa = 8.5TLVVSFATPGFKK233 pKa = 10.49EE234 pKa = 3.91GDD236 pKa = 3.24IKK238 pKa = 11.29LSFLNGNTTVRR249 pKa = 11.84SILEE253 pKa = 4.55DD254 pKa = 2.4IWRR257 pKa = 11.84AMGRR261 pKa = 11.84WAYY264 pKa = 9.68ISSSYY269 pKa = 10.55HH270 pKa = 5.45VDD272 pKa = 4.22GIVVV276 pKa = 3.73
MM1 pKa = 7.79KK2 pKa = 9.17KK3 pKa = 7.62TQQHH7 pKa = 5.05SLRR10 pKa = 11.84PIMDD14 pKa = 3.79EE15 pKa = 3.9LNEE18 pKa = 4.09KK19 pKa = 10.13FKK21 pKa = 11.35QFLDD25 pKa = 3.65DD26 pKa = 3.95GARR29 pKa = 11.84QNILSDD35 pKa = 3.37EE36 pKa = 4.63FKK38 pKa = 10.85RR39 pKa = 11.84DD40 pKa = 3.08VNAILGEE47 pKa = 4.18MEE49 pKa = 4.55DD50 pKa = 4.19EE51 pKa = 4.39EE52 pKa = 5.05MPLAPNDD59 pKa = 3.8HH60 pKa = 6.38SPVRR64 pKa = 11.84QSTSLYY70 pKa = 10.93SPDD73 pKa = 4.62HH74 pKa = 7.08DD75 pKa = 5.13LDD77 pKa = 3.98TYY79 pKa = 10.9TDD81 pKa = 4.42PGCQSPPGSVCDD93 pKa = 4.13PLDD96 pKa = 5.06DD97 pKa = 4.23EE98 pKa = 5.11ACSILGEE105 pKa = 4.12LEE107 pKa = 4.47RR108 pKa = 11.84IVTLSDD114 pKa = 4.7PGNQWEE120 pKa = 4.62GASHH124 pKa = 6.92IVWTSPGVYY133 pKa = 8.46STGRR137 pKa = 11.84EE138 pKa = 3.82FQQAKK143 pKa = 8.5TLLKK147 pKa = 10.57SVLRR151 pKa = 11.84SMNRR155 pKa = 11.84DD156 pKa = 2.86SGLNYY161 pKa = 9.8VLNVGKK167 pKa = 9.58PDD169 pKa = 3.41TLFITRR175 pKa = 11.84PDD177 pKa = 3.48SLTVARR183 pKa = 11.84NVRR186 pKa = 11.84PKK188 pKa = 10.16IPEE191 pKa = 3.83NLFVDD196 pKa = 3.43NDD198 pKa = 2.84KK199 pKa = 11.22WEE201 pKa = 4.75FIKK204 pKa = 11.03AQFASGIEE212 pKa = 4.09LKK214 pKa = 10.96DD215 pKa = 3.09VDD217 pKa = 4.6EE218 pKa = 4.9PGKK221 pKa = 8.5TLVVSFATPGFKK233 pKa = 10.49EE234 pKa = 3.91GDD236 pKa = 3.24IKK238 pKa = 11.29LSFLNGNTTVRR249 pKa = 11.84SILEE253 pKa = 4.55DD254 pKa = 2.4IWRR257 pKa = 11.84AMGRR261 pKa = 11.84WAYY264 pKa = 9.68ISSSYY269 pKa = 10.55HH270 pKa = 5.45VDD272 pKa = 4.22GIVVV276 pKa = 3.73
Molecular weight: 30.95 kDa
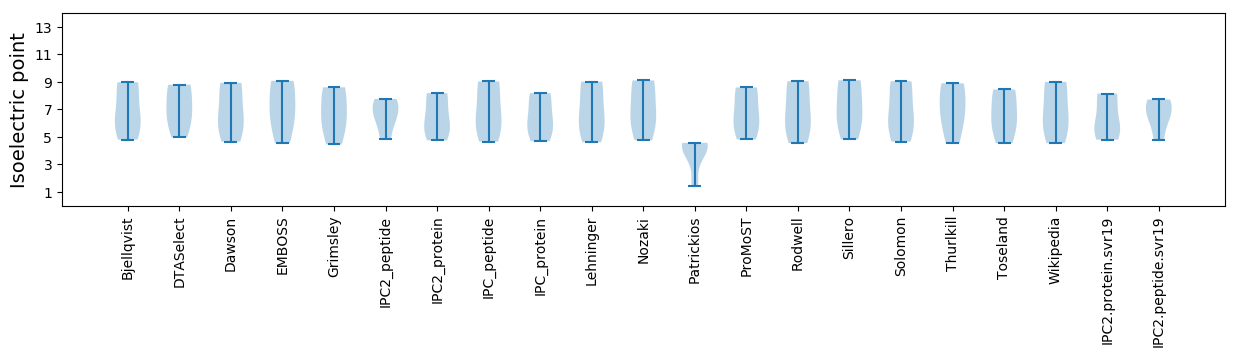
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A336USW8|A0A336USW8_9RHAB Matrix protein OS=Gata virus OX=1911435 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 9.22KK3 pKa = 8.4TRR5 pKa = 11.84TPNRR9 pKa = 11.84TEE11 pKa = 3.48MLSRR15 pKa = 11.84FRR17 pKa = 11.84SKK19 pKa = 11.24NKK21 pKa = 10.6DD22 pKa = 3.01NDD24 pKa = 4.13AGSSDD29 pKa = 3.78YY30 pKa = 9.9WANPPNKK37 pKa = 9.05NPEE40 pKa = 3.74MSMPNSPVYY49 pKa = 10.92SFTGSWAISGRR60 pKa = 11.84FEE62 pKa = 3.92IRR64 pKa = 11.84TKK66 pKa = 10.68IPIQSGAKK74 pKa = 10.21LLDD77 pKa = 5.24LIGAWLDD84 pKa = 4.05GYY86 pKa = 10.67SGSYY90 pKa = 9.63SFHH93 pKa = 7.0GIFTGLYY100 pKa = 9.96LLMALHH106 pKa = 6.81SLPSQEE112 pKa = 3.89NQTYY116 pKa = 10.06VYY118 pKa = 10.93SLDD121 pKa = 3.29LSKK124 pKa = 10.84RR125 pKa = 11.84IAIQVTRR132 pKa = 11.84QDD134 pKa = 4.08VINQEE139 pKa = 4.58ADD141 pKa = 3.72LSWVNQEE148 pKa = 3.8GTGLEE153 pKa = 3.75WCVIKK158 pKa = 10.18FKK160 pKa = 10.65IHH162 pKa = 6.57LKK164 pKa = 7.28PTRR167 pKa = 11.84RR168 pKa = 11.84MCIPFADD175 pKa = 4.64AYY177 pKa = 8.42KK178 pKa = 10.6QPMHH182 pKa = 6.84TGADD186 pKa = 3.52PPPFEE191 pKa = 3.99EE192 pKa = 5.24VYY194 pKa = 10.66FRR196 pKa = 11.84FWNPQSGYY204 pKa = 10.67CSIIHH209 pKa = 6.53
MM1 pKa = 7.58KK2 pKa = 9.22KK3 pKa = 8.4TRR5 pKa = 11.84TPNRR9 pKa = 11.84TEE11 pKa = 3.48MLSRR15 pKa = 11.84FRR17 pKa = 11.84SKK19 pKa = 11.24NKK21 pKa = 10.6DD22 pKa = 3.01NDD24 pKa = 4.13AGSSDD29 pKa = 3.78YY30 pKa = 9.9WANPPNKK37 pKa = 9.05NPEE40 pKa = 3.74MSMPNSPVYY49 pKa = 10.92SFTGSWAISGRR60 pKa = 11.84FEE62 pKa = 3.92IRR64 pKa = 11.84TKK66 pKa = 10.68IPIQSGAKK74 pKa = 10.21LLDD77 pKa = 5.24LIGAWLDD84 pKa = 4.05GYY86 pKa = 10.67SGSYY90 pKa = 9.63SFHH93 pKa = 7.0GIFTGLYY100 pKa = 9.96LLMALHH106 pKa = 6.81SLPSQEE112 pKa = 3.89NQTYY116 pKa = 10.06VYY118 pKa = 10.93SLDD121 pKa = 3.29LSKK124 pKa = 10.84RR125 pKa = 11.84IAIQVTRR132 pKa = 11.84QDD134 pKa = 4.08VINQEE139 pKa = 4.58ADD141 pKa = 3.72LSWVNQEE148 pKa = 3.8GTGLEE153 pKa = 3.75WCVIKK158 pKa = 10.18FKK160 pKa = 10.65IHH162 pKa = 6.57LKK164 pKa = 7.28PTRR167 pKa = 11.84RR168 pKa = 11.84MCIPFADD175 pKa = 4.64AYY177 pKa = 8.42KK178 pKa = 10.6QPMHH182 pKa = 6.84TGADD186 pKa = 3.52PPPFEE191 pKa = 3.99EE192 pKa = 5.24VYY194 pKa = 10.66FRR196 pKa = 11.84FWNPQSGYY204 pKa = 10.67CSIIHH209 pKa = 6.53
Molecular weight: 23.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3581 |
209 |
2135 |
716.2 |
81.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.937 ± 0.682 | 1.759 ± 0.504 |
5.892 ± 0.475 | 4.971 ± 0.23 |
4.524 ± 0.289 | 5.781 ± 0.154 |
2.457 ± 0.317 | 6.898 ± 0.449 |
6.171 ± 0.329 | 9.969 ± 1.135 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 0.303 | 5.334 ± 0.4 |
4.775 ± 0.6 | 3.239 ± 0.207 |
5.166 ± 0.456 | 9.69 ± 0.254 |
5.613 ± 0.473 | 5.781 ± 0.476 |
1.787 ± 0.146 | 3.658 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
