
Xingshan nematode virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.91
Get precalculated fractions of proteins
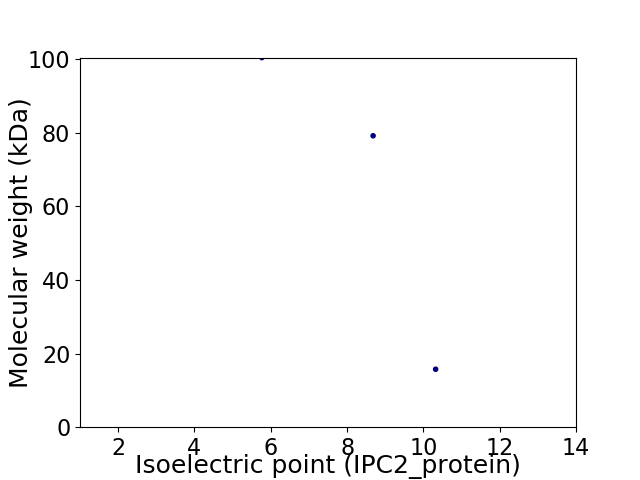
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFD6|A0A1L3KFD6_9VIRU RdRp catalytic domain-containing protein OS=Xingshan nematode virus 6 OX=1923765 PE=4 SV=1
MM1 pKa = 7.66ASDD4 pKa = 3.96SLKK7 pKa = 9.56STDD10 pKa = 3.28YY11 pKa = 10.98VGRR14 pKa = 11.84LRR16 pKa = 11.84SALGNRR22 pKa = 11.84RR23 pKa = 11.84TVQDD27 pKa = 3.85RR28 pKa = 11.84EE29 pKa = 4.41TEE31 pKa = 4.17DD32 pKa = 3.3FTVASGLAFYY42 pKa = 10.64SALFTPVATSILPGDD57 pKa = 3.96VRR59 pKa = 11.84EE60 pKa = 4.56AMCGLNTVSQKK71 pKa = 9.82TSGSGCFSLHH81 pKa = 5.83HH82 pKa = 5.96TVSDD86 pKa = 3.52IMPLNLEE93 pKa = 4.74LGDD96 pKa = 3.76EE97 pKa = 4.36KK98 pKa = 11.05KK99 pKa = 11.18GNNRR103 pKa = 11.84AARR106 pKa = 11.84KK107 pKa = 9.7VSGGNGKK114 pKa = 8.67VKK116 pKa = 10.41RR117 pKa = 11.84KK118 pKa = 10.58SNDD121 pKa = 3.07QGWDD125 pKa = 3.25VKK127 pKa = 10.84NQAVHH132 pKa = 5.11GRR134 pKa = 11.84EE135 pKa = 4.2VKK137 pKa = 10.52VSTGLGCHH145 pKa = 5.75QGTPPVTSLYY155 pKa = 10.03AQLVHH160 pKa = 6.42RR161 pKa = 11.84QCLPLEE167 pKa = 4.67AGSGTQASTTNNPVYY182 pKa = 10.31TDD184 pKa = 3.66YY185 pKa = 11.72GSLAFMTEE193 pKa = 4.13VVTGQPGSWKK203 pKa = 10.32DD204 pKa = 3.34DD205 pKa = 3.62APILRR210 pKa = 11.84MLLLGDD216 pKa = 4.03IFAMADD222 pKa = 3.64RR223 pKa = 11.84YY224 pKa = 10.9KK225 pKa = 10.8LDD227 pKa = 4.91PIWYY231 pKa = 9.54KK232 pKa = 10.41LARR235 pKa = 11.84DD236 pKa = 4.19ANPSSIEE243 pKa = 3.51WHH245 pKa = 5.01MTGLPKK251 pKa = 10.67EE252 pKa = 4.34GGVNSRR258 pKa = 11.84SVQITAVTLDD268 pKa = 2.84VWAALRR274 pKa = 11.84VGSLKK279 pKa = 10.9DD280 pKa = 3.35STLSEE285 pKa = 4.14LVQGVNCAVIPISSNKK301 pKa = 9.5LDD303 pKa = 4.12EE304 pKa = 4.3AWIWLYY310 pKa = 10.96LAAFCTSRR318 pKa = 11.84IWNGLHH324 pKa = 4.62QWEE327 pKa = 4.67VSLDD331 pKa = 3.37PSVRR335 pKa = 11.84SKK337 pKa = 11.07SEE339 pKa = 3.81KK340 pKa = 9.97LLEE343 pKa = 4.6TIFTTRR349 pKa = 11.84SSLCMVEE356 pKa = 3.77GHH358 pKa = 5.8EE359 pKa = 5.3RR360 pKa = 11.84ILLVLTDD367 pKa = 3.33VTSRR371 pKa = 11.84LGYY374 pKa = 8.45KK375 pKa = 6.51TTRR378 pKa = 11.84LARR381 pKa = 11.84EE382 pKa = 4.07KK383 pKa = 10.24IVPIWQKK390 pKa = 9.64TDD392 pKa = 3.44PKK394 pKa = 10.29VGWVEE399 pKa = 4.08IIDD402 pKa = 3.82EE403 pKa = 4.24LRR405 pKa = 11.84YY406 pKa = 8.84ATGVTTLDD414 pKa = 3.76YY415 pKa = 11.08PHH417 pKa = 7.33LGDD420 pKa = 4.24LWRR423 pKa = 11.84MAWKK427 pKa = 10.67EE428 pKa = 3.84MMASVATGDD437 pKa = 3.74SVNRR441 pKa = 11.84CMALVAEE448 pKa = 5.16ATRR451 pKa = 11.84VMDD454 pKa = 3.13VGFYY458 pKa = 10.66SGGEE462 pKa = 4.14VNVMSLGKK470 pKa = 9.96IEE472 pKa = 4.89GLPEE476 pKa = 4.68LDD478 pKa = 3.04GVVWKK483 pKa = 10.42NRR485 pKa = 11.84QWEE488 pKa = 4.58SMTADD493 pKa = 3.47DD494 pKa = 6.02AKK496 pKa = 11.02FGLQDD501 pKa = 4.16ASLSWVSVSGMRR513 pKa = 11.84PDD515 pKa = 4.24FAWTVLDD522 pKa = 4.0PEE524 pKa = 5.08KK525 pKa = 10.52GRR527 pKa = 11.84SMLTVGGIDD536 pKa = 3.09ASLYY540 pKa = 9.16CHH542 pKa = 6.67EE543 pKa = 4.82ATGDD547 pKa = 3.34MRR549 pKa = 11.84MARR552 pKa = 11.84YY553 pKa = 9.83VGIISGAEE561 pKa = 3.43RR562 pKa = 11.84VALEE566 pKa = 3.89TCVAVSEE573 pKa = 4.33NVSAIASFMACAVGWMQSVAGIPWYY598 pKa = 10.37KK599 pKa = 9.14WAHH602 pKa = 5.09HH603 pKa = 6.67HH604 pKa = 6.22NLPEE608 pKa = 4.12LTSIDD613 pKa = 4.2DD614 pKa = 4.3LASMMLDD621 pKa = 3.34AASDD625 pKa = 3.57GHH627 pKa = 6.17IKK629 pKa = 10.32RR630 pKa = 11.84SYY632 pKa = 11.29ADD634 pKa = 3.71PTWCTWVEE642 pKa = 3.58NNIFHH647 pKa = 6.27GQPWSKK653 pKa = 10.77VCTTILPWWAMKK665 pKa = 9.7FVIQKK670 pKa = 9.9VGYY673 pKa = 8.62HH674 pKa = 6.53LDD676 pKa = 3.26VGLVEE681 pKa = 4.11RR682 pKa = 11.84QKK684 pKa = 10.59PVEE687 pKa = 4.15GKK689 pKa = 10.17RR690 pKa = 11.84PRR692 pKa = 11.84IGYY695 pKa = 6.04WATMASEE702 pKa = 4.21WQNVARR708 pKa = 11.84VTSRR712 pKa = 11.84ITVRR716 pKa = 11.84YY717 pKa = 8.4PDD719 pKa = 3.68EE720 pKa = 4.18CVEE723 pKa = 4.64YY724 pKa = 9.55YY725 pKa = 10.06TSSYY729 pKa = 11.83DD730 pKa = 3.79NANLGAGVVGYY741 pKa = 9.59NWHH744 pKa = 6.6RR745 pKa = 11.84AAAEE749 pKa = 4.02YY750 pKa = 11.13GEE752 pKa = 4.69SQYY755 pKa = 11.13HH756 pKa = 5.09WQTSARR762 pKa = 11.84PWNKK766 pKa = 10.09SVDD769 pKa = 3.71SSTLITPVSYY779 pKa = 11.4AMVYY783 pKa = 10.22GNPSGMLITMTRR795 pKa = 11.84NRR797 pKa = 11.84VGSGMDD803 pKa = 3.52DD804 pKa = 3.19QDD806 pKa = 3.74VTGRR810 pKa = 11.84WPDD813 pKa = 3.46PWWNWLLEE821 pKa = 4.19GVYY824 pKa = 9.95EE825 pKa = 4.35ALPHH829 pKa = 6.68LLQGNLLGAALAGMSGGVNAFFRR852 pKa = 11.84EE853 pKa = 3.84RR854 pKa = 11.84LNPVSAQKK862 pKa = 9.93EE863 pKa = 3.93QQARR867 pKa = 11.84MEE869 pKa = 4.37SAARR873 pKa = 11.84AADD876 pKa = 4.11KK877 pKa = 10.8ILTAGKK883 pKa = 10.18DD884 pKa = 3.55LVEE887 pKa = 4.66HH888 pKa = 6.45GKK890 pKa = 9.91AAAAEE895 pKa = 4.25TVAAARR901 pKa = 11.84EE902 pKa = 4.24FAGEE906 pKa = 4.23AGPSGPAA913 pKa = 3.07
MM1 pKa = 7.66ASDD4 pKa = 3.96SLKK7 pKa = 9.56STDD10 pKa = 3.28YY11 pKa = 10.98VGRR14 pKa = 11.84LRR16 pKa = 11.84SALGNRR22 pKa = 11.84RR23 pKa = 11.84TVQDD27 pKa = 3.85RR28 pKa = 11.84EE29 pKa = 4.41TEE31 pKa = 4.17DD32 pKa = 3.3FTVASGLAFYY42 pKa = 10.64SALFTPVATSILPGDD57 pKa = 3.96VRR59 pKa = 11.84EE60 pKa = 4.56AMCGLNTVSQKK71 pKa = 9.82TSGSGCFSLHH81 pKa = 5.83HH82 pKa = 5.96TVSDD86 pKa = 3.52IMPLNLEE93 pKa = 4.74LGDD96 pKa = 3.76EE97 pKa = 4.36KK98 pKa = 11.05KK99 pKa = 11.18GNNRR103 pKa = 11.84AARR106 pKa = 11.84KK107 pKa = 9.7VSGGNGKK114 pKa = 8.67VKK116 pKa = 10.41RR117 pKa = 11.84KK118 pKa = 10.58SNDD121 pKa = 3.07QGWDD125 pKa = 3.25VKK127 pKa = 10.84NQAVHH132 pKa = 5.11GRR134 pKa = 11.84EE135 pKa = 4.2VKK137 pKa = 10.52VSTGLGCHH145 pKa = 5.75QGTPPVTSLYY155 pKa = 10.03AQLVHH160 pKa = 6.42RR161 pKa = 11.84QCLPLEE167 pKa = 4.67AGSGTQASTTNNPVYY182 pKa = 10.31TDD184 pKa = 3.66YY185 pKa = 11.72GSLAFMTEE193 pKa = 4.13VVTGQPGSWKK203 pKa = 10.32DD204 pKa = 3.34DD205 pKa = 3.62APILRR210 pKa = 11.84MLLLGDD216 pKa = 4.03IFAMADD222 pKa = 3.64RR223 pKa = 11.84YY224 pKa = 10.9KK225 pKa = 10.8LDD227 pKa = 4.91PIWYY231 pKa = 9.54KK232 pKa = 10.41LARR235 pKa = 11.84DD236 pKa = 4.19ANPSSIEE243 pKa = 3.51WHH245 pKa = 5.01MTGLPKK251 pKa = 10.67EE252 pKa = 4.34GGVNSRR258 pKa = 11.84SVQITAVTLDD268 pKa = 2.84VWAALRR274 pKa = 11.84VGSLKK279 pKa = 10.9DD280 pKa = 3.35STLSEE285 pKa = 4.14LVQGVNCAVIPISSNKK301 pKa = 9.5LDD303 pKa = 4.12EE304 pKa = 4.3AWIWLYY310 pKa = 10.96LAAFCTSRR318 pKa = 11.84IWNGLHH324 pKa = 4.62QWEE327 pKa = 4.67VSLDD331 pKa = 3.37PSVRR335 pKa = 11.84SKK337 pKa = 11.07SEE339 pKa = 3.81KK340 pKa = 9.97LLEE343 pKa = 4.6TIFTTRR349 pKa = 11.84SSLCMVEE356 pKa = 3.77GHH358 pKa = 5.8EE359 pKa = 5.3RR360 pKa = 11.84ILLVLTDD367 pKa = 3.33VTSRR371 pKa = 11.84LGYY374 pKa = 8.45KK375 pKa = 6.51TTRR378 pKa = 11.84LARR381 pKa = 11.84EE382 pKa = 4.07KK383 pKa = 10.24IVPIWQKK390 pKa = 9.64TDD392 pKa = 3.44PKK394 pKa = 10.29VGWVEE399 pKa = 4.08IIDD402 pKa = 3.82EE403 pKa = 4.24LRR405 pKa = 11.84YY406 pKa = 8.84ATGVTTLDD414 pKa = 3.76YY415 pKa = 11.08PHH417 pKa = 7.33LGDD420 pKa = 4.24LWRR423 pKa = 11.84MAWKK427 pKa = 10.67EE428 pKa = 3.84MMASVATGDD437 pKa = 3.74SVNRR441 pKa = 11.84CMALVAEE448 pKa = 5.16ATRR451 pKa = 11.84VMDD454 pKa = 3.13VGFYY458 pKa = 10.66SGGEE462 pKa = 4.14VNVMSLGKK470 pKa = 9.96IEE472 pKa = 4.89GLPEE476 pKa = 4.68LDD478 pKa = 3.04GVVWKK483 pKa = 10.42NRR485 pKa = 11.84QWEE488 pKa = 4.58SMTADD493 pKa = 3.47DD494 pKa = 6.02AKK496 pKa = 11.02FGLQDD501 pKa = 4.16ASLSWVSVSGMRR513 pKa = 11.84PDD515 pKa = 4.24FAWTVLDD522 pKa = 4.0PEE524 pKa = 5.08KK525 pKa = 10.52GRR527 pKa = 11.84SMLTVGGIDD536 pKa = 3.09ASLYY540 pKa = 9.16CHH542 pKa = 6.67EE543 pKa = 4.82ATGDD547 pKa = 3.34MRR549 pKa = 11.84MARR552 pKa = 11.84YY553 pKa = 9.83VGIISGAEE561 pKa = 3.43RR562 pKa = 11.84VALEE566 pKa = 3.89TCVAVSEE573 pKa = 4.33NVSAIASFMACAVGWMQSVAGIPWYY598 pKa = 10.37KK599 pKa = 9.14WAHH602 pKa = 5.09HH603 pKa = 6.67HH604 pKa = 6.22NLPEE608 pKa = 4.12LTSIDD613 pKa = 4.2DD614 pKa = 4.3LASMMLDD621 pKa = 3.34AASDD625 pKa = 3.57GHH627 pKa = 6.17IKK629 pKa = 10.32RR630 pKa = 11.84SYY632 pKa = 11.29ADD634 pKa = 3.71PTWCTWVEE642 pKa = 3.58NNIFHH647 pKa = 6.27GQPWSKK653 pKa = 10.77VCTTILPWWAMKK665 pKa = 9.7FVIQKK670 pKa = 9.9VGYY673 pKa = 8.62HH674 pKa = 6.53LDD676 pKa = 3.26VGLVEE681 pKa = 4.11RR682 pKa = 11.84QKK684 pKa = 10.59PVEE687 pKa = 4.15GKK689 pKa = 10.17RR690 pKa = 11.84PRR692 pKa = 11.84IGYY695 pKa = 6.04WATMASEE702 pKa = 4.21WQNVARR708 pKa = 11.84VTSRR712 pKa = 11.84ITVRR716 pKa = 11.84YY717 pKa = 8.4PDD719 pKa = 3.68EE720 pKa = 4.18CVEE723 pKa = 4.64YY724 pKa = 9.55YY725 pKa = 10.06TSSYY729 pKa = 11.83DD730 pKa = 3.79NANLGAGVVGYY741 pKa = 9.59NWHH744 pKa = 6.6RR745 pKa = 11.84AAAEE749 pKa = 4.02YY750 pKa = 11.13GEE752 pKa = 4.69SQYY755 pKa = 11.13HH756 pKa = 5.09WQTSARR762 pKa = 11.84PWNKK766 pKa = 10.09SVDD769 pKa = 3.71SSTLITPVSYY779 pKa = 11.4AMVYY783 pKa = 10.22GNPSGMLITMTRR795 pKa = 11.84NRR797 pKa = 11.84VGSGMDD803 pKa = 3.52DD804 pKa = 3.19QDD806 pKa = 3.74VTGRR810 pKa = 11.84WPDD813 pKa = 3.46PWWNWLLEE821 pKa = 4.19GVYY824 pKa = 9.95EE825 pKa = 4.35ALPHH829 pKa = 6.68LLQGNLLGAALAGMSGGVNAFFRR852 pKa = 11.84EE853 pKa = 3.84RR854 pKa = 11.84LNPVSAQKK862 pKa = 9.93EE863 pKa = 3.93QQARR867 pKa = 11.84MEE869 pKa = 4.37SAARR873 pKa = 11.84AADD876 pKa = 4.11KK877 pKa = 10.8ILTAGKK883 pKa = 10.18DD884 pKa = 3.55LVEE887 pKa = 4.66HH888 pKa = 6.45GKK890 pKa = 9.91AAAAEE895 pKa = 4.25TVAAARR901 pKa = 11.84EE902 pKa = 4.24FAGEE906 pKa = 4.23AGPSGPAA913 pKa = 3.07
Molecular weight: 100.45 kDa
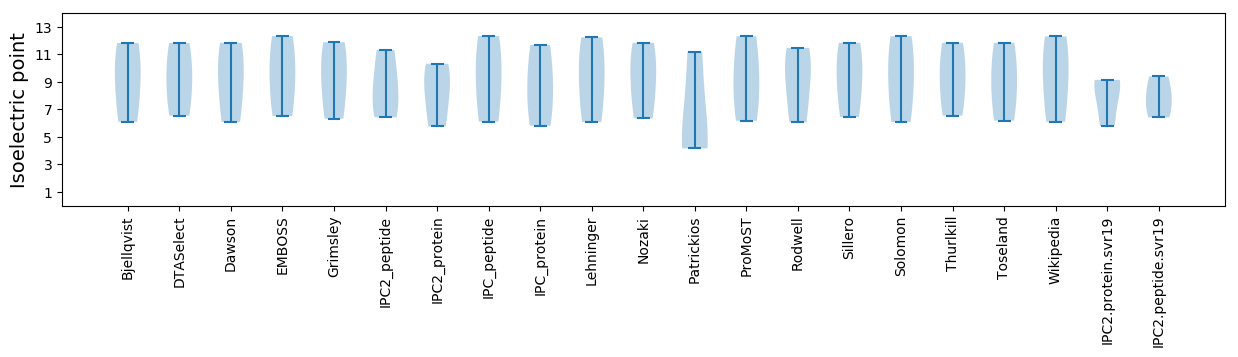
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFL3|A0A1L3KFL3_9VIRU Uncharacterized protein OS=Xingshan nematode virus 6 OX=1923765 PE=4 SV=1
MM1 pKa = 7.18FSYY4 pKa = 10.65VRR6 pKa = 11.84RR7 pKa = 11.84GPRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GTEE16 pKa = 3.55TASAPLVVWGLRR28 pKa = 11.84LIRR31 pKa = 11.84RR32 pKa = 11.84NSIPLAGWGLPGYY45 pKa = 10.39SGVLTRR51 pKa = 11.84NLGTTSSINGGRR63 pKa = 11.84FCMVRR68 pKa = 11.84DD69 pKa = 4.19LRR71 pKa = 11.84GSWLLVVGCKK81 pKa = 9.84QPVKK85 pKa = 9.73QAHH88 pKa = 6.17RR89 pKa = 11.84STWEE93 pKa = 3.64CSGALGVTGNNKK105 pKa = 9.09ARR107 pKa = 11.84RR108 pKa = 11.84VYY110 pKa = 10.86RR111 pKa = 11.84MAGISGRR118 pKa = 11.84IPPRR122 pKa = 11.84PLPLGVPCHH131 pKa = 5.41FVVRR135 pKa = 11.84VHH137 pKa = 6.18GCPWHH142 pKa = 5.83GG143 pKa = 3.35
MM1 pKa = 7.18FSYY4 pKa = 10.65VRR6 pKa = 11.84RR7 pKa = 11.84GPRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GTEE16 pKa = 3.55TASAPLVVWGLRR28 pKa = 11.84LIRR31 pKa = 11.84RR32 pKa = 11.84NSIPLAGWGLPGYY45 pKa = 10.39SGVLTRR51 pKa = 11.84NLGTTSSINGGRR63 pKa = 11.84FCMVRR68 pKa = 11.84DD69 pKa = 4.19LRR71 pKa = 11.84GSWLLVVGCKK81 pKa = 9.84QPVKK85 pKa = 9.73QAHH88 pKa = 6.17RR89 pKa = 11.84STWEE93 pKa = 3.64CSGALGVTGNNKK105 pKa = 9.09ARR107 pKa = 11.84RR108 pKa = 11.84VYY110 pKa = 10.86RR111 pKa = 11.84MAGISGRR118 pKa = 11.84IPPRR122 pKa = 11.84PLPLGVPCHH131 pKa = 5.41FVVRR135 pKa = 11.84VHH137 pKa = 6.18GCPWHH142 pKa = 5.83GG143 pKa = 3.35
Molecular weight: 15.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1746 |
143 |
913 |
582.0 |
65.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.362 ± 1.131 | 1.947 ± 0.459 |
5.04 ± 1.112 | 4.811 ± 0.904 |
2.176 ± 0.242 | 8.534 ± 1.403 |
2.234 ± 0.144 | 3.608 ± 0.028 |
4.353 ± 0.6 | 9.737 ± 1.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.207 ± 0.296 | 2.749 ± 0.715 |
4.525 ± 0.831 | 3.036 ± 0.44 |
7.388 ± 2.015 | 7.102 ± 0.751 |
5.613 ± 0.553 | 8.247 ± 0.495 |
4.066 ± 0.321 | 3.265 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
