
Thermobifida phage P318
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
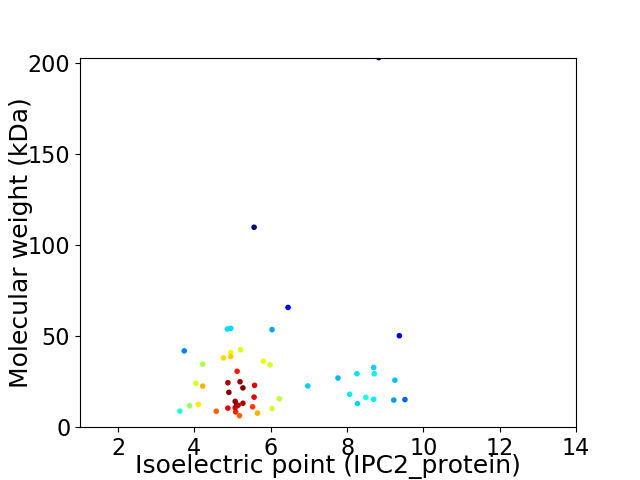
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3T0ID44|A0A3T0ID44_9CAUD FAD-dependent thymidylate synthase OS=Thermobifida phage P318 OX=2500138 PE=3 SV=1
MM1 pKa = 7.12STTTTSYY8 pKa = 10.97GHH10 pKa = 5.82WTNIVDD16 pKa = 4.31HH17 pKa = 6.65KK18 pKa = 11.1ASTVGQTVLRR28 pKa = 11.84FLTAADD34 pKa = 4.07PDD36 pKa = 3.74WAEE39 pKa = 3.89EE40 pKa = 4.14LEE42 pKa = 4.43EE43 pKa = 4.58NGVLEE48 pKa = 5.66DD49 pKa = 4.5IIEE52 pKa = 4.32EE53 pKa = 3.96YY54 pKa = 10.93RR55 pKa = 11.84NAINDD60 pKa = 4.03ALPAGIVLAGSEE72 pKa = 4.47FFGPVDD78 pKa = 4.23PDD80 pKa = 3.51PAEE83 pKa = 3.81WDD85 pKa = 2.74GDD87 pKa = 4.23YY88 pKa = 11.08PVTSDD93 pKa = 3.74GRR95 pKa = 11.84LDD97 pKa = 3.13IAAIVAEE104 pKa = 4.17VDD106 pKa = 3.19LGEE109 pKa = 4.33IVAKK113 pKa = 10.29FDD115 pKa = 3.59EE116 pKa = 4.73VDD118 pKa = 3.2PDD120 pKa = 4.08GYY122 pKa = 10.47VAAVATSADD131 pKa = 3.39VVVGNNCVVAVFEE144 pKa = 4.42RR145 pKa = 11.84QVTGYY150 pKa = 10.64RR151 pKa = 11.84SGEE154 pKa = 4.2DD155 pKa = 3.56GEE157 pKa = 4.38EE158 pKa = 4.14TPVYY162 pKa = 10.53SLGDD166 pKa = 3.36IVYY169 pKa = 9.67EE170 pKa = 4.3ADD172 pKa = 2.92TGIPMDD178 pKa = 5.17ADD180 pKa = 3.82DD181 pKa = 4.09ATTRR185 pKa = 11.84ATAAATEE192 pKa = 4.14LLEE195 pKa = 4.16EE196 pKa = 4.74AGWEE200 pKa = 4.3VVSEE204 pKa = 4.04WEE206 pKa = 4.0YY207 pKa = 11.47SDD209 pKa = 3.46NSVYY213 pKa = 11.37AHH215 pKa = 6.57ATRR218 pKa = 11.84HH219 pKa = 5.62NPNQYY224 pKa = 9.05TYY226 pKa = 10.7AIEE229 pKa = 4.02TSVDD233 pKa = 3.41GEE235 pKa = 4.37SWSVEE240 pKa = 4.37VISDD244 pKa = 3.54AQTAEE249 pKa = 4.22TPEE252 pKa = 4.08EE253 pKa = 4.17CASRR257 pKa = 11.84LLVDD261 pKa = 3.47WLGEE265 pKa = 4.13FEE267 pKa = 4.48EE268 pKa = 5.06TPQYY272 pKa = 10.3SRR274 pKa = 11.84CVVWEE279 pKa = 4.08GNIDD283 pKa = 4.11DD284 pKa = 4.27VDD286 pKa = 3.61KK287 pKa = 11.08AAAIAYY293 pKa = 7.92PPDD296 pKa = 4.0SYY298 pKa = 11.33PVTVYY303 pKa = 9.6WSIVRR308 pKa = 11.84GEE310 pKa = 4.25APYY313 pKa = 10.68AAAAEE318 pKa = 4.37PGPLGIPVPDD328 pKa = 3.59AVLEE332 pKa = 4.21LGARR336 pKa = 11.84FGLGQDD342 pKa = 3.13PDD344 pKa = 4.35VYY346 pKa = 11.62VMVAPAEE353 pKa = 4.01LAGPIPGEE361 pKa = 3.51LAYY364 pKa = 10.93YY365 pKa = 10.49RR366 pKa = 11.84GTLPYY371 pKa = 9.66ATALQVWAEE380 pKa = 3.77IQEE383 pKa = 4.22RR384 pKa = 11.84QEE386 pKa = 3.82RR387 pKa = 3.86
MM1 pKa = 7.12STTTTSYY8 pKa = 10.97GHH10 pKa = 5.82WTNIVDD16 pKa = 4.31HH17 pKa = 6.65KK18 pKa = 11.1ASTVGQTVLRR28 pKa = 11.84FLTAADD34 pKa = 4.07PDD36 pKa = 3.74WAEE39 pKa = 3.89EE40 pKa = 4.14LEE42 pKa = 4.43EE43 pKa = 4.58NGVLEE48 pKa = 5.66DD49 pKa = 4.5IIEE52 pKa = 4.32EE53 pKa = 3.96YY54 pKa = 10.93RR55 pKa = 11.84NAINDD60 pKa = 4.03ALPAGIVLAGSEE72 pKa = 4.47FFGPVDD78 pKa = 4.23PDD80 pKa = 3.51PAEE83 pKa = 3.81WDD85 pKa = 2.74GDD87 pKa = 4.23YY88 pKa = 11.08PVTSDD93 pKa = 3.74GRR95 pKa = 11.84LDD97 pKa = 3.13IAAIVAEE104 pKa = 4.17VDD106 pKa = 3.19LGEE109 pKa = 4.33IVAKK113 pKa = 10.29FDD115 pKa = 3.59EE116 pKa = 4.73VDD118 pKa = 3.2PDD120 pKa = 4.08GYY122 pKa = 10.47VAAVATSADD131 pKa = 3.39VVVGNNCVVAVFEE144 pKa = 4.42RR145 pKa = 11.84QVTGYY150 pKa = 10.64RR151 pKa = 11.84SGEE154 pKa = 4.2DD155 pKa = 3.56GEE157 pKa = 4.38EE158 pKa = 4.14TPVYY162 pKa = 10.53SLGDD166 pKa = 3.36IVYY169 pKa = 9.67EE170 pKa = 4.3ADD172 pKa = 2.92TGIPMDD178 pKa = 5.17ADD180 pKa = 3.82DD181 pKa = 4.09ATTRR185 pKa = 11.84ATAAATEE192 pKa = 4.14LLEE195 pKa = 4.16EE196 pKa = 4.74AGWEE200 pKa = 4.3VVSEE204 pKa = 4.04WEE206 pKa = 4.0YY207 pKa = 11.47SDD209 pKa = 3.46NSVYY213 pKa = 11.37AHH215 pKa = 6.57ATRR218 pKa = 11.84HH219 pKa = 5.62NPNQYY224 pKa = 9.05TYY226 pKa = 10.7AIEE229 pKa = 4.02TSVDD233 pKa = 3.41GEE235 pKa = 4.37SWSVEE240 pKa = 4.37VISDD244 pKa = 3.54AQTAEE249 pKa = 4.22TPEE252 pKa = 4.08EE253 pKa = 4.17CASRR257 pKa = 11.84LLVDD261 pKa = 3.47WLGEE265 pKa = 4.13FEE267 pKa = 4.48EE268 pKa = 5.06TPQYY272 pKa = 10.3SRR274 pKa = 11.84CVVWEE279 pKa = 4.08GNIDD283 pKa = 4.11DD284 pKa = 4.27VDD286 pKa = 3.61KK287 pKa = 11.08AAAIAYY293 pKa = 7.92PPDD296 pKa = 4.0SYY298 pKa = 11.33PVTVYY303 pKa = 9.6WSIVRR308 pKa = 11.84GEE310 pKa = 4.25APYY313 pKa = 10.68AAAAEE318 pKa = 4.37PGPLGIPVPDD328 pKa = 3.59AVLEE332 pKa = 4.21LGARR336 pKa = 11.84FGLGQDD342 pKa = 3.13PDD344 pKa = 4.35VYY346 pKa = 11.62VMVAPAEE353 pKa = 4.01LAGPIPGEE361 pKa = 3.51LAYY364 pKa = 10.93YY365 pKa = 10.49RR366 pKa = 11.84GTLPYY371 pKa = 9.66ATALQVWAEE380 pKa = 3.77IQEE383 pKa = 4.22RR384 pKa = 11.84QEE386 pKa = 3.82RR387 pKa = 3.86
Molecular weight: 41.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9R573|A0A3Q9R573_9CAUD Minor tail protein OS=Thermobifida phage P318 OX=2500138 PE=4 SV=1
MM1 pKa = 7.22KK2 pKa = 10.12HH3 pKa = 4.6SHH5 pKa = 5.02QPIRR9 pKa = 11.84KK10 pKa = 8.24ILRR13 pKa = 11.84NSLPVVEE20 pKa = 5.4KK21 pKa = 10.98VDD23 pKa = 4.43DD24 pKa = 4.17LFFVSALEE32 pKa = 3.91IPRR35 pKa = 11.84NVVDD39 pKa = 4.37RR40 pKa = 11.84LNHH43 pKa = 5.62VSDD46 pKa = 4.52DD47 pKa = 3.84ASSGRR52 pKa = 11.84QIIHH56 pKa = 7.08ANRR59 pKa = 11.84SAPARR64 pKa = 11.84QCIQGRR70 pKa = 11.84QPVGVRR76 pKa = 11.84DD77 pKa = 3.61RR78 pKa = 11.84PGGKK82 pKa = 9.25RR83 pKa = 11.84FNRR86 pKa = 11.84LGRR89 pKa = 11.84QDD91 pKa = 4.23KK92 pKa = 10.86IVDD95 pKa = 3.86FVVNHH100 pKa = 6.14IQPIVFGLLGQGDD113 pKa = 4.86YY114 pKa = 8.91PTEE117 pKa = 4.38IGGEE121 pKa = 4.01INSLLKK127 pKa = 10.7HH128 pKa = 6.13GSCPFQLLTGQISAEE143 pKa = 4.22LKK145 pKa = 10.55PLTPLYY151 pKa = 10.3KK152 pKa = 10.23NPKK155 pKa = 7.87HH156 pKa = 6.34AVGGTPARR164 pKa = 11.84VRR166 pKa = 11.84LGQASLQLCWSDD178 pKa = 3.26VTNDD182 pKa = 2.98SDD184 pKa = 3.8VWGVHH189 pKa = 5.38HH190 pKa = 7.09NRR192 pKa = 11.84LFLSNVRR199 pKa = 11.84KK200 pKa = 10.03ASSFPCFSQVLAQRR214 pKa = 11.84RR215 pKa = 11.84IHH217 pKa = 6.23PVSAKK222 pKa = 8.39YY223 pKa = 9.75RR224 pKa = 11.84RR225 pKa = 11.84HH226 pKa = 6.34FGHH229 pKa = 7.18DD230 pKa = 3.06HH231 pKa = 6.03QSRR234 pKa = 11.84KK235 pKa = 9.74SGKK238 pKa = 9.58KK239 pKa = 9.4PGDD242 pKa = 3.25VFLTVFVFLLHH253 pKa = 6.49HH254 pKa = 6.75PIVLQRR260 pKa = 11.84PTVQISHH267 pKa = 5.54EE268 pKa = 4.06TSPRR272 pKa = 11.84VGGGNEE278 pKa = 3.67VTSPVKK284 pKa = 10.5QVGNRR289 pKa = 11.84LFHH292 pKa = 6.04HH293 pKa = 6.94HH294 pKa = 6.82SGIPCGSTFPANARR308 pKa = 11.84IASQDD313 pKa = 3.32RR314 pKa = 11.84PNGQLLVRR322 pKa = 11.84VTNLDD327 pKa = 2.78GRR329 pKa = 11.84LRR331 pKa = 11.84PNSDD335 pKa = 4.18RR336 pKa = 11.84IHH338 pKa = 5.91GHH340 pKa = 5.56QGHH343 pKa = 6.82EE344 pKa = 4.11DD345 pKa = 3.5ADD347 pKa = 4.13RR348 pKa = 11.84VGLSVRR354 pKa = 11.84VQNVDD359 pKa = 3.28NVSKK363 pKa = 10.72RR364 pKa = 11.84AVEE367 pKa = 3.93NFFARR372 pKa = 11.84DD373 pKa = 3.33GTFNHH378 pKa = 6.92CYY380 pKa = 9.99LRR382 pKa = 11.84SSLFYY387 pKa = 9.26QAKK390 pKa = 7.41WNYY393 pKa = 8.18WINGEE398 pKa = 4.29PPTVCRR404 pKa = 11.84GLSGLCQVTVPDD416 pKa = 4.04VPGFPPTLAAPWRR429 pKa = 11.84TAEE432 pKa = 4.35CPGNKK437 pKa = 9.42LARR440 pKa = 11.84QGSDD444 pKa = 2.71RR445 pKa = 11.84PAPPARR451 pKa = 11.84PP452 pKa = 3.42
MM1 pKa = 7.22KK2 pKa = 10.12HH3 pKa = 4.6SHH5 pKa = 5.02QPIRR9 pKa = 11.84KK10 pKa = 8.24ILRR13 pKa = 11.84NSLPVVEE20 pKa = 5.4KK21 pKa = 10.98VDD23 pKa = 4.43DD24 pKa = 4.17LFFVSALEE32 pKa = 3.91IPRR35 pKa = 11.84NVVDD39 pKa = 4.37RR40 pKa = 11.84LNHH43 pKa = 5.62VSDD46 pKa = 4.52DD47 pKa = 3.84ASSGRR52 pKa = 11.84QIIHH56 pKa = 7.08ANRR59 pKa = 11.84SAPARR64 pKa = 11.84QCIQGRR70 pKa = 11.84QPVGVRR76 pKa = 11.84DD77 pKa = 3.61RR78 pKa = 11.84PGGKK82 pKa = 9.25RR83 pKa = 11.84FNRR86 pKa = 11.84LGRR89 pKa = 11.84QDD91 pKa = 4.23KK92 pKa = 10.86IVDD95 pKa = 3.86FVVNHH100 pKa = 6.14IQPIVFGLLGQGDD113 pKa = 4.86YY114 pKa = 8.91PTEE117 pKa = 4.38IGGEE121 pKa = 4.01INSLLKK127 pKa = 10.7HH128 pKa = 6.13GSCPFQLLTGQISAEE143 pKa = 4.22LKK145 pKa = 10.55PLTPLYY151 pKa = 10.3KK152 pKa = 10.23NPKK155 pKa = 7.87HH156 pKa = 6.34AVGGTPARR164 pKa = 11.84VRR166 pKa = 11.84LGQASLQLCWSDD178 pKa = 3.26VTNDD182 pKa = 2.98SDD184 pKa = 3.8VWGVHH189 pKa = 5.38HH190 pKa = 7.09NRR192 pKa = 11.84LFLSNVRR199 pKa = 11.84KK200 pKa = 10.03ASSFPCFSQVLAQRR214 pKa = 11.84RR215 pKa = 11.84IHH217 pKa = 6.23PVSAKK222 pKa = 8.39YY223 pKa = 9.75RR224 pKa = 11.84RR225 pKa = 11.84HH226 pKa = 6.34FGHH229 pKa = 7.18DD230 pKa = 3.06HH231 pKa = 6.03QSRR234 pKa = 11.84KK235 pKa = 9.74SGKK238 pKa = 9.58KK239 pKa = 9.4PGDD242 pKa = 3.25VFLTVFVFLLHH253 pKa = 6.49HH254 pKa = 6.75PIVLQRR260 pKa = 11.84PTVQISHH267 pKa = 5.54EE268 pKa = 4.06TSPRR272 pKa = 11.84VGGGNEE278 pKa = 3.67VTSPVKK284 pKa = 10.5QVGNRR289 pKa = 11.84LFHH292 pKa = 6.04HH293 pKa = 6.94HH294 pKa = 6.82SGIPCGSTFPANARR308 pKa = 11.84IASQDD313 pKa = 3.32RR314 pKa = 11.84PNGQLLVRR322 pKa = 11.84VTNLDD327 pKa = 2.78GRR329 pKa = 11.84LRR331 pKa = 11.84PNSDD335 pKa = 4.18RR336 pKa = 11.84IHH338 pKa = 5.91GHH340 pKa = 5.56QGHH343 pKa = 6.82EE344 pKa = 4.11DD345 pKa = 3.5ADD347 pKa = 4.13RR348 pKa = 11.84VGLSVRR354 pKa = 11.84VQNVDD359 pKa = 3.28NVSKK363 pKa = 10.72RR364 pKa = 11.84AVEE367 pKa = 3.93NFFARR372 pKa = 11.84DD373 pKa = 3.33GTFNHH378 pKa = 6.92CYY380 pKa = 9.99LRR382 pKa = 11.84SSLFYY387 pKa = 9.26QAKK390 pKa = 7.41WNYY393 pKa = 8.18WINGEE398 pKa = 4.29PPTVCRR404 pKa = 11.84GLSGLCQVTVPDD416 pKa = 4.04VPGFPPTLAAPWRR429 pKa = 11.84TAEE432 pKa = 4.35CPGNKK437 pKa = 9.42LARR440 pKa = 11.84QGSDD444 pKa = 2.71RR445 pKa = 11.84PAPPARR451 pKa = 11.84PP452 pKa = 3.42
Molecular weight: 50.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13907 |
62 |
1899 |
267.4 |
29.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.405 ± 0.598 | 0.748 ± 0.126 |
5.947 ± 0.388 | 7.313 ± 0.388 |
3.3 ± 0.124 | 8.262 ± 0.372 |
1.826 ± 0.218 | 5.242 ± 0.238 |
4.473 ± 0.34 | 7.644 ± 0.265 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.1 ± 0.138 | 3.574 ± 0.164 |
5.228 ± 0.292 | 3.056 ± 0.21 |
6.996 ± 0.286 | 6.285 ± 0.206 |
5.544 ± 0.262 | 8.046 ± 0.289 |
1.769 ± 0.126 | 3.243 ± 0.306 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
