
Chaetoceros tenuissimus DNA virus type-II
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.9
Get precalculated fractions of proteins
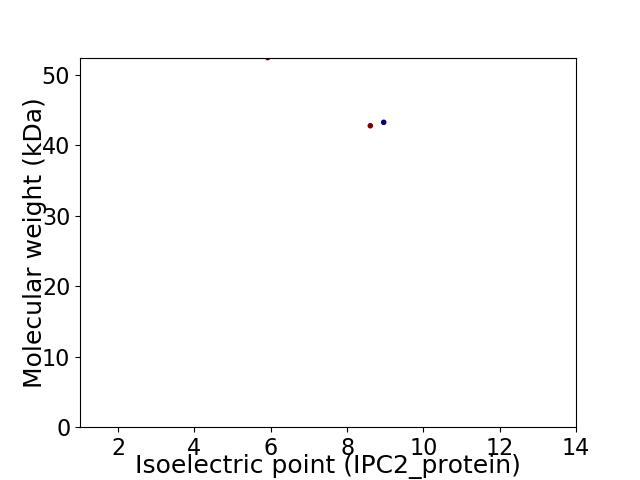
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B6VL42|A0A0B6VL42_9VIRU Uncharacterized protein OS=Chaetoceros tenuissimus DNA virus type-II OX=1516127 PE=4 SV=1
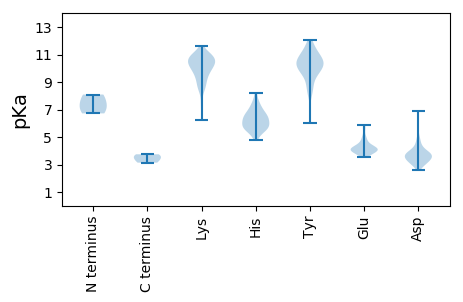
MM1 pKa = 7.34VFRR4 pKa = 11.84TMSDD8 pKa = 2.61NSFDD12 pKa = 3.54IQQIICDD19 pKa = 4.11EE20 pKa = 4.16ASIDD24 pKa = 3.83CSITIDD30 pKa = 3.75TQPVDD35 pKa = 4.25DD36 pKa = 5.39LSASTEE42 pKa = 3.83YY43 pKa = 11.03RR44 pKa = 11.84EE45 pKa = 4.1VSNNTRR51 pKa = 11.84IGRR54 pKa = 11.84AILTYY59 pKa = 9.81FPPTSEE65 pKa = 4.17SKK67 pKa = 10.28YY68 pKa = 10.51LEE70 pKa = 4.19PEE72 pKa = 4.81FILGRR77 pKa = 11.84ANVHH81 pKa = 6.35RR82 pKa = 11.84VSNWFSQYY90 pKa = 8.92EE91 pKa = 4.14LCPEE95 pKa = 4.18TEE97 pKa = 4.08RR98 pKa = 11.84KK99 pKa = 9.65HH100 pKa = 5.06IHH102 pKa = 6.38AYY104 pKa = 10.62VEE106 pKa = 4.96FKK108 pKa = 10.95HH109 pKa = 5.91SVRR112 pKa = 11.84PFFLTLDD119 pKa = 3.9TLFKK123 pKa = 10.94SHH125 pKa = 7.69DD126 pKa = 4.26LGWDD130 pKa = 3.03CRR132 pKa = 11.84TARR135 pKa = 11.84KK136 pKa = 7.62TSKK139 pKa = 10.25DD140 pKa = 3.42QRR142 pKa = 11.84QGAINYY148 pKa = 6.03VHH150 pKa = 7.01KK151 pKa = 10.9LEE153 pKa = 4.29SRR155 pKa = 11.84MDD157 pKa = 3.56GFDD160 pKa = 3.61PYY162 pKa = 10.09TWPGNEE168 pKa = 4.39DD169 pKa = 3.46KK170 pKa = 10.89DD171 pKa = 4.61LKK173 pKa = 10.64FDD175 pKa = 3.85PSFVRR180 pKa = 11.84GNRR183 pKa = 11.84KK184 pKa = 8.87KK185 pKa = 10.62DD186 pKa = 3.18KK187 pKa = 10.83RR188 pKa = 11.84KK189 pKa = 10.3ARR191 pKa = 11.84DD192 pKa = 3.38DD193 pKa = 4.5AIEE196 pKa = 3.9EE197 pKa = 3.69QRR199 pKa = 11.84QYY201 pKa = 11.01IEE203 pKa = 4.12SKK205 pKa = 9.72PRR207 pKa = 11.84IWTWDD212 pKa = 3.48QIVHH216 pKa = 6.07EE217 pKa = 4.58SEE219 pKa = 4.04EE220 pKa = 4.54SKK222 pKa = 11.11KK223 pKa = 10.96LLVTCSWGKK232 pKa = 9.75KK233 pKa = 6.24YY234 pKa = 10.62HH235 pKa = 6.38EE236 pKa = 4.89GRR238 pKa = 11.84HH239 pKa = 5.65AEE241 pKa = 4.14DD242 pKa = 3.29KK243 pKa = 11.0RR244 pKa = 11.84RR245 pKa = 11.84DD246 pKa = 3.52VTDD249 pKa = 3.59VIIYY253 pKa = 9.63YY254 pKa = 10.43GAGGTGKK261 pKa = 7.4TTMCMEE267 pKa = 4.92HH268 pKa = 7.44DD269 pKa = 4.07STSSLEE275 pKa = 4.1PRR277 pKa = 11.84VEE279 pKa = 3.56RR280 pKa = 11.84YY281 pKa = 8.52YY282 pKa = 10.96RR283 pKa = 11.84RR284 pKa = 11.84NPDD287 pKa = 4.5DD288 pKa = 4.03GNFWGGGRR296 pKa = 11.84TAYY299 pKa = 9.76KK300 pKa = 8.82GQRR303 pKa = 11.84VIHH306 pKa = 5.98FEE308 pKa = 3.94EE309 pKa = 5.09FGGQEE314 pKa = 3.6PFSRR318 pKa = 11.84LKK320 pKa = 10.11EE321 pKa = 3.9VCDD324 pKa = 3.38IGKK327 pKa = 9.09PGPSVNIKK335 pKa = 10.38GAGVDD340 pKa = 4.22LNHH343 pKa = 6.56HH344 pKa = 5.34TVIFSSNIHH353 pKa = 5.46PAGWYY358 pKa = 8.22RR359 pKa = 11.84HH360 pKa = 5.44LWKK363 pKa = 10.38DD364 pKa = 3.92DD365 pKa = 3.65PKK367 pKa = 10.84QFHH370 pKa = 6.3PFWRR374 pKa = 11.84RR375 pKa = 11.84VTQVKK380 pKa = 8.48FFPSHH385 pKa = 6.74RR386 pKa = 11.84PDD388 pKa = 3.07GTLNIPDD395 pKa = 4.93EE396 pKa = 4.56DD397 pKa = 4.34TPPYY401 pKa = 10.44IIDD404 pKa = 3.56QTDD407 pKa = 2.76EE408 pKa = 4.02WKK410 pKa = 10.77EE411 pKa = 3.69FAGDD415 pKa = 3.4YY416 pKa = 9.62SKK418 pKa = 11.65CMDD421 pKa = 4.39HH422 pKa = 8.2ADD424 pKa = 3.15IHH426 pKa = 5.74WPVRR430 pKa = 11.84EE431 pKa = 3.92VAVEE435 pKa = 4.03GFSASFGHH443 pKa = 5.68SHH445 pKa = 4.8QTQRR449 pKa = 11.84SS450 pKa = 3.49
MM1 pKa = 7.34VFRR4 pKa = 11.84TMSDD8 pKa = 2.61NSFDD12 pKa = 3.54IQQIICDD19 pKa = 4.11EE20 pKa = 4.16ASIDD24 pKa = 3.83CSITIDD30 pKa = 3.75TQPVDD35 pKa = 4.25DD36 pKa = 5.39LSASTEE42 pKa = 3.83YY43 pKa = 11.03RR44 pKa = 11.84EE45 pKa = 4.1VSNNTRR51 pKa = 11.84IGRR54 pKa = 11.84AILTYY59 pKa = 9.81FPPTSEE65 pKa = 4.17SKK67 pKa = 10.28YY68 pKa = 10.51LEE70 pKa = 4.19PEE72 pKa = 4.81FILGRR77 pKa = 11.84ANVHH81 pKa = 6.35RR82 pKa = 11.84VSNWFSQYY90 pKa = 8.92EE91 pKa = 4.14LCPEE95 pKa = 4.18TEE97 pKa = 4.08RR98 pKa = 11.84KK99 pKa = 9.65HH100 pKa = 5.06IHH102 pKa = 6.38AYY104 pKa = 10.62VEE106 pKa = 4.96FKK108 pKa = 10.95HH109 pKa = 5.91SVRR112 pKa = 11.84PFFLTLDD119 pKa = 3.9TLFKK123 pKa = 10.94SHH125 pKa = 7.69DD126 pKa = 4.26LGWDD130 pKa = 3.03CRR132 pKa = 11.84TARR135 pKa = 11.84KK136 pKa = 7.62TSKK139 pKa = 10.25DD140 pKa = 3.42QRR142 pKa = 11.84QGAINYY148 pKa = 6.03VHH150 pKa = 7.01KK151 pKa = 10.9LEE153 pKa = 4.29SRR155 pKa = 11.84MDD157 pKa = 3.56GFDD160 pKa = 3.61PYY162 pKa = 10.09TWPGNEE168 pKa = 4.39DD169 pKa = 3.46KK170 pKa = 10.89DD171 pKa = 4.61LKK173 pKa = 10.64FDD175 pKa = 3.85PSFVRR180 pKa = 11.84GNRR183 pKa = 11.84KK184 pKa = 8.87KK185 pKa = 10.62DD186 pKa = 3.18KK187 pKa = 10.83RR188 pKa = 11.84KK189 pKa = 10.3ARR191 pKa = 11.84DD192 pKa = 3.38DD193 pKa = 4.5AIEE196 pKa = 3.9EE197 pKa = 3.69QRR199 pKa = 11.84QYY201 pKa = 11.01IEE203 pKa = 4.12SKK205 pKa = 9.72PRR207 pKa = 11.84IWTWDD212 pKa = 3.48QIVHH216 pKa = 6.07EE217 pKa = 4.58SEE219 pKa = 4.04EE220 pKa = 4.54SKK222 pKa = 11.11KK223 pKa = 10.96LLVTCSWGKK232 pKa = 9.75KK233 pKa = 6.24YY234 pKa = 10.62HH235 pKa = 6.38EE236 pKa = 4.89GRR238 pKa = 11.84HH239 pKa = 5.65AEE241 pKa = 4.14DD242 pKa = 3.29KK243 pKa = 11.0RR244 pKa = 11.84RR245 pKa = 11.84DD246 pKa = 3.52VTDD249 pKa = 3.59VIIYY253 pKa = 9.63YY254 pKa = 10.43GAGGTGKK261 pKa = 7.4TTMCMEE267 pKa = 4.92HH268 pKa = 7.44DD269 pKa = 4.07STSSLEE275 pKa = 4.1PRR277 pKa = 11.84VEE279 pKa = 3.56RR280 pKa = 11.84YY281 pKa = 8.52YY282 pKa = 10.96RR283 pKa = 11.84RR284 pKa = 11.84NPDD287 pKa = 4.5DD288 pKa = 4.03GNFWGGGRR296 pKa = 11.84TAYY299 pKa = 9.76KK300 pKa = 8.82GQRR303 pKa = 11.84VIHH306 pKa = 5.98FEE308 pKa = 3.94EE309 pKa = 5.09FGGQEE314 pKa = 3.6PFSRR318 pKa = 11.84LKK320 pKa = 10.11EE321 pKa = 3.9VCDD324 pKa = 3.38IGKK327 pKa = 9.09PGPSVNIKK335 pKa = 10.38GAGVDD340 pKa = 4.22LNHH343 pKa = 6.56HH344 pKa = 5.34TVIFSSNIHH353 pKa = 5.46PAGWYY358 pKa = 8.22RR359 pKa = 11.84HH360 pKa = 5.44LWKK363 pKa = 10.38DD364 pKa = 3.92DD365 pKa = 3.65PKK367 pKa = 10.84QFHH370 pKa = 6.3PFWRR374 pKa = 11.84RR375 pKa = 11.84VTQVKK380 pKa = 8.48FFPSHH385 pKa = 6.74RR386 pKa = 11.84PDD388 pKa = 3.07GTLNIPDD395 pKa = 4.93EE396 pKa = 4.56DD397 pKa = 4.34TPPYY401 pKa = 10.44IIDD404 pKa = 3.56QTDD407 pKa = 2.76EE408 pKa = 4.02WKK410 pKa = 10.77EE411 pKa = 3.69FAGDD415 pKa = 3.4YY416 pKa = 9.62SKK418 pKa = 11.65CMDD421 pKa = 4.39HH422 pKa = 8.2ADD424 pKa = 3.15IHH426 pKa = 5.74WPVRR430 pKa = 11.84EE431 pKa = 3.92VAVEE435 pKa = 4.03GFSASFGHH443 pKa = 5.68SHH445 pKa = 4.8QTQRR449 pKa = 11.84SS450 pKa = 3.49
Molecular weight: 52.46 kDa
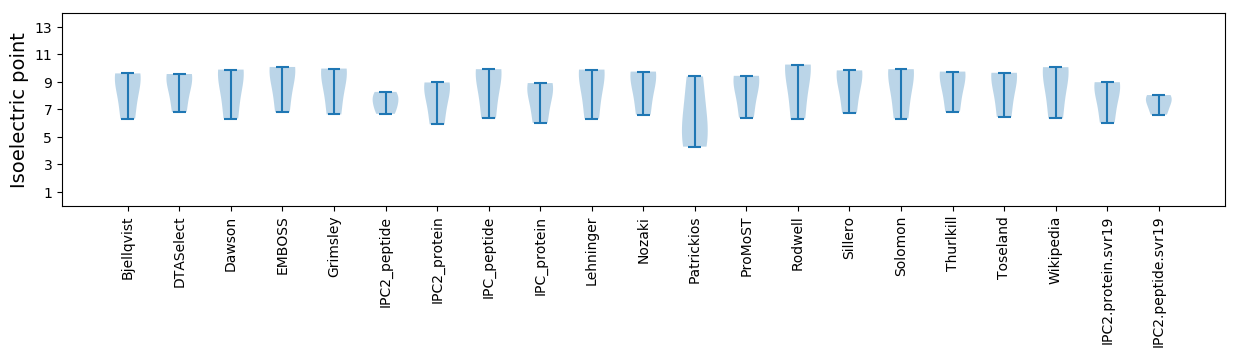
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B6VL42|A0A0B6VL42_9VIRU Uncharacterized protein OS=Chaetoceros tenuissimus DNA virus type-II OX=1516127 PE=4 SV=1
MM1 pKa = 6.72VRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84GGKK8 pKa = 6.81TAGSKK13 pKa = 9.5RR14 pKa = 11.84PKK16 pKa = 9.54MSSKK20 pKa = 10.78NFGANRR26 pKa = 11.84KK27 pKa = 9.23RR28 pKa = 11.84DD29 pKa = 3.58FRR31 pKa = 11.84RR32 pKa = 11.84PARR35 pKa = 11.84KK36 pKa = 9.57SKK38 pKa = 10.47AKK40 pKa = 9.87KK41 pKa = 9.05ARR43 pKa = 11.84SMAPAKK49 pKa = 8.89TVRR52 pKa = 11.84KK53 pKa = 8.32STTAGAHH60 pKa = 5.57SKK62 pKa = 10.22HH63 pKa = 6.23FSVIGNPFSKK73 pKa = 9.75ATQQPQIPDD82 pKa = 2.99GRR84 pKa = 11.84MLEE87 pKa = 4.2SLPRR91 pKa = 11.84RR92 pKa = 11.84CQLVTEE98 pKa = 4.4IRR100 pKa = 11.84NNVTVGSNPTYY111 pKa = 10.63ILVAPSLGLAFQAYY125 pKa = 9.86QDD127 pKa = 3.89TNVPGGLDD135 pKa = 3.21SSVYY139 pKa = 9.73GLQNRR144 pKa = 11.84GCTVRR149 pKa = 11.84ANLSATSIEE158 pKa = 4.31NYY160 pKa = 10.47NDD162 pKa = 3.07IAKK165 pKa = 9.27WRR167 pKa = 11.84IVSQGINLKK176 pKa = 10.42LNNVEE181 pKa = 4.74DD182 pKa = 4.89EE183 pKa = 4.05NDD185 pKa = 3.08GWYY188 pKa = 8.29EE189 pKa = 3.82ACRR192 pKa = 11.84FQHH195 pKa = 6.89DD196 pKa = 3.7WTPDD200 pKa = 3.54EE201 pKa = 4.29LCLRR205 pKa = 11.84STEE208 pKa = 4.22NDD210 pKa = 2.89ASTISQDD217 pKa = 2.91EE218 pKa = 4.35DD219 pKa = 3.75LVMGVISSSFMNGALNTIGNNMVEE243 pKa = 4.01QRR245 pKa = 11.84GYY247 pKa = 11.32EE248 pKa = 3.98SGLLKK253 pKa = 10.6NIHH256 pKa = 6.07KK257 pKa = 10.72RR258 pKa = 11.84MFQLHH263 pKa = 6.56NNTSAIRR270 pKa = 11.84PKK272 pKa = 9.88TLQGQFNYY280 pKa = 10.37GSEE283 pKa = 3.95ITFSGTEE290 pKa = 3.81SEE292 pKa = 5.26ARR294 pKa = 11.84FTDD297 pKa = 3.87VPSNRR302 pKa = 11.84QLVDD306 pKa = 3.89SLWHH310 pKa = 6.55NDD312 pKa = 3.33YY313 pKa = 11.57DD314 pKa = 4.92CILIKK319 pKa = 10.49LYY321 pKa = 10.22PRR323 pKa = 11.84EE324 pKa = 4.06NTGAAGQTGSALIVNAIQNLEE345 pKa = 4.11LQYY348 pKa = 11.37SPTSDD353 pKa = 2.86LSTYY357 pKa = 10.42HH358 pKa = 6.83IANKK362 pKa = 8.51RR363 pKa = 11.84ARR365 pKa = 11.84MVEE368 pKa = 3.73AKK370 pKa = 10.21LDD372 pKa = 3.76KK373 pKa = 11.13KK374 pKa = 11.09NNTDD378 pKa = 3.22AAGEE382 pKa = 4.09PFVPGSSRR390 pKa = 3.14
MM1 pKa = 6.72VRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84GGKK8 pKa = 6.81TAGSKK13 pKa = 9.5RR14 pKa = 11.84PKK16 pKa = 9.54MSSKK20 pKa = 10.78NFGANRR26 pKa = 11.84KK27 pKa = 9.23RR28 pKa = 11.84DD29 pKa = 3.58FRR31 pKa = 11.84RR32 pKa = 11.84PARR35 pKa = 11.84KK36 pKa = 9.57SKK38 pKa = 10.47AKK40 pKa = 9.87KK41 pKa = 9.05ARR43 pKa = 11.84SMAPAKK49 pKa = 8.89TVRR52 pKa = 11.84KK53 pKa = 8.32STTAGAHH60 pKa = 5.57SKK62 pKa = 10.22HH63 pKa = 6.23FSVIGNPFSKK73 pKa = 9.75ATQQPQIPDD82 pKa = 2.99GRR84 pKa = 11.84MLEE87 pKa = 4.2SLPRR91 pKa = 11.84RR92 pKa = 11.84CQLVTEE98 pKa = 4.4IRR100 pKa = 11.84NNVTVGSNPTYY111 pKa = 10.63ILVAPSLGLAFQAYY125 pKa = 9.86QDD127 pKa = 3.89TNVPGGLDD135 pKa = 3.21SSVYY139 pKa = 9.73GLQNRR144 pKa = 11.84GCTVRR149 pKa = 11.84ANLSATSIEE158 pKa = 4.31NYY160 pKa = 10.47NDD162 pKa = 3.07IAKK165 pKa = 9.27WRR167 pKa = 11.84IVSQGINLKK176 pKa = 10.42LNNVEE181 pKa = 4.74DD182 pKa = 4.89EE183 pKa = 4.05NDD185 pKa = 3.08GWYY188 pKa = 8.29EE189 pKa = 3.82ACRR192 pKa = 11.84FQHH195 pKa = 6.89DD196 pKa = 3.7WTPDD200 pKa = 3.54EE201 pKa = 4.29LCLRR205 pKa = 11.84STEE208 pKa = 4.22NDD210 pKa = 2.89ASTISQDD217 pKa = 2.91EE218 pKa = 4.35DD219 pKa = 3.75LVMGVISSSFMNGALNTIGNNMVEE243 pKa = 4.01QRR245 pKa = 11.84GYY247 pKa = 11.32EE248 pKa = 3.98SGLLKK253 pKa = 10.6NIHH256 pKa = 6.07KK257 pKa = 10.72RR258 pKa = 11.84MFQLHH263 pKa = 6.56NNTSAIRR270 pKa = 11.84PKK272 pKa = 9.88TLQGQFNYY280 pKa = 10.37GSEE283 pKa = 3.95ITFSGTEE290 pKa = 3.81SEE292 pKa = 5.26ARR294 pKa = 11.84FTDD297 pKa = 3.87VPSNRR302 pKa = 11.84QLVDD306 pKa = 3.89SLWHH310 pKa = 6.55NDD312 pKa = 3.33YY313 pKa = 11.57DD314 pKa = 4.92CILIKK319 pKa = 10.49LYY321 pKa = 10.22PRR323 pKa = 11.84EE324 pKa = 4.06NTGAAGQTGSALIVNAIQNLEE345 pKa = 4.11LQYY348 pKa = 11.37SPTSDD353 pKa = 2.86LSTYY357 pKa = 10.42HH358 pKa = 6.83IANKK362 pKa = 8.51RR363 pKa = 11.84ARR365 pKa = 11.84MVEE368 pKa = 3.73AKK370 pKa = 10.21LDD372 pKa = 3.76KK373 pKa = 11.13KK374 pKa = 11.09NNTDD378 pKa = 3.22AAGEE382 pKa = 4.09PFVPGSSRR390 pKa = 3.14
Molecular weight: 43.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1226 |
386 |
450 |
408.7 |
46.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.444 ± 1.045 | 1.06 ± 0.408 |
7.341 ± 0.868 | 5.791 ± 0.581 |
4.323 ± 0.5 | 7.586 ± 0.663 |
3.263 ± 0.646 | 5.465 ± 0.211 |
7.993 ± 1.407 | 5.628 ± 0.728 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.039 ± 0.308 | 4.812 ± 1.199 |
4.16 ± 0.625 | 3.752 ± 0.314 |
6.607 ± 0.703 | 7.912 ± 0.463 |
5.628 ± 0.441 | 5.546 ± 0.242 |
1.631 ± 0.441 | 3.018 ± 0.342 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
