
Piscine myocarditis-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 8.01
Get precalculated fractions of proteins
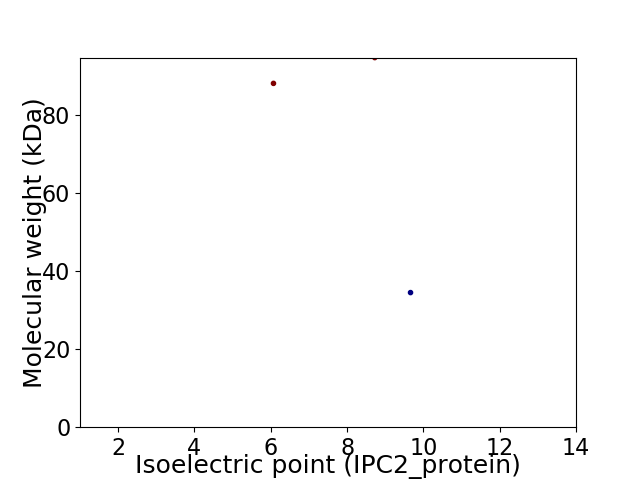
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A125SJD2|A0A125SJD2_9VIRU RNA-directed RNA polymerase OS=Piscine myocarditis-like virus OX=1798085 PE=3 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84IAGDD6 pKa = 3.59RR7 pKa = 11.84QTVLLGLPSGKK18 pKa = 10.44GGISVLVGDD27 pKa = 4.92FATNLEE33 pKa = 4.39SVARR37 pKa = 11.84AIFTDD42 pKa = 3.25TNMVVDD48 pKa = 3.85ATIEE52 pKa = 3.82DD53 pKa = 3.73RR54 pKa = 11.84ARR56 pKa = 11.84ARR58 pKa = 11.84NHH60 pKa = 5.73TILRR64 pKa = 11.84EE65 pKa = 3.96GLIDD69 pKa = 3.29SRR71 pKa = 11.84MYY73 pKa = 10.1MSAGRR78 pKa = 11.84TTQVALWAAGPPVAVGVNPTWYY100 pKa = 9.41ILRR103 pKa = 11.84PGKK106 pKa = 10.53VEE108 pKa = 5.32GIMLTQSSSMASLAPLFRR126 pKa = 11.84GVEE129 pKa = 4.07LGPFASAVRR138 pKa = 11.84ADD140 pKa = 3.33VGFYY144 pKa = 11.11SMNVVLGALRR154 pKa = 11.84AAGFNAEE161 pKa = 4.06NGRR164 pKa = 11.84VSFVEE169 pKa = 3.52PLLRR173 pKa = 11.84VQCKK177 pKa = 9.93ILDD180 pKa = 4.03GQDD183 pKa = 3.3RR184 pKa = 11.84GEE186 pKa = 4.13SPYY189 pKa = 10.91SIVGGLTSQLVNPVTQRR206 pKa = 11.84VWGNFFPGGPNQAAWVAADD225 pKa = 3.54TVSRR229 pKa = 11.84VVNIEE234 pKa = 3.59DD235 pKa = 4.31FMRR238 pKa = 11.84EE239 pKa = 3.85AQGEE243 pKa = 4.2NRR245 pKa = 11.84FAAGWGPGFWNKK257 pKa = 9.26MGATGVAVVPIKK269 pKa = 10.24TSGAMLAQQNSAWTLAHH286 pKa = 6.09MEE288 pKa = 4.15YY289 pKa = 9.73PRR291 pKa = 11.84KK292 pKa = 9.76VRR294 pKa = 11.84QVDD297 pKa = 3.69TNVISINAALAAVGAEE313 pKa = 3.66RR314 pKa = 11.84EE315 pKa = 4.29YY316 pKa = 11.22TNTSCTHH323 pKa = 4.78VPGPYY328 pKa = 10.11GKK330 pKa = 10.0VLFVIVDD337 pKa = 3.51QTTDD341 pKa = 2.92RR342 pKa = 11.84AVQVAVEE349 pKa = 4.23GAVGVLLTTPALHH362 pKa = 5.68YY363 pKa = 10.58VIGGIDD369 pKa = 3.4MGIGLMVTRR378 pKa = 11.84EE379 pKa = 3.76VGLAIDD385 pKa = 3.68QAALEE390 pKa = 4.89NIMAMEE396 pKa = 4.43RR397 pKa = 11.84WVEE400 pKa = 4.04FYY402 pKa = 11.31GNEE405 pKa = 4.14EE406 pKa = 4.21DD407 pKa = 3.64WQSATTIVGCAYY419 pKa = 9.21VTFGVEE425 pKa = 4.15MRR427 pKa = 11.84RR428 pKa = 11.84HH429 pKa = 4.81TNRR432 pKa = 11.84VEE434 pKa = 3.95GGYY437 pKa = 9.44YY438 pKa = 8.16WQVGANPPFPLGGLANWTRR457 pKa = 11.84LDD459 pKa = 3.77NLTAAQQAAIYY470 pKa = 10.76ADD472 pKa = 3.14IQNTVEE478 pKa = 4.15APWGGFRR485 pKa = 11.84VRR487 pKa = 11.84NPVPTAAPNAMYY499 pKa = 9.96TRR501 pKa = 11.84FEE503 pKa = 4.06PVLAVGTVAMLWRR516 pKa = 11.84PEE518 pKa = 4.05HH519 pKa = 6.85PMLAPLPDD527 pKa = 4.19RR528 pKa = 11.84PSALADD534 pKa = 2.85ICYY537 pKa = 10.13QRR539 pKa = 11.84GRR541 pKa = 11.84MCAAQEE547 pKa = 4.46DD548 pKa = 4.94IVVGQLGFPEE558 pKa = 4.25DD559 pKa = 4.03MIRR562 pKa = 11.84APVGVGNVGLAQRR575 pKa = 11.84LIRR578 pKa = 11.84KK579 pKa = 6.56MCNVNNQILEE589 pKa = 4.27AKK591 pKa = 9.46HH592 pKa = 6.02ASTVHH597 pKa = 5.22VVLNGYY603 pKa = 8.67WSIGAGVTYY612 pKa = 10.28RR613 pKa = 11.84AIYY616 pKa = 9.01AQSGPGTMRR625 pKa = 11.84KK626 pKa = 9.71RR627 pKa = 11.84IPWAEE632 pKa = 3.87MRR634 pKa = 11.84SLGFDD639 pKa = 3.36ADD641 pKa = 4.05LVKK644 pKa = 10.94NDD646 pKa = 4.2LYY648 pKa = 11.46GIGSGDD654 pKa = 3.46FVAVNATIGALGGIWDD670 pKa = 3.95VVRR673 pKa = 11.84YY674 pKa = 7.15PLPVPSKK681 pKa = 9.12EE682 pKa = 3.71ARR684 pKa = 11.84AAMNTTNCMGGWTRR698 pKa = 11.84AVTRR702 pKa = 11.84VDD704 pKa = 3.05NLGTIRR710 pKa = 11.84EE711 pKa = 4.46GGLEE715 pKa = 4.14PEE717 pKa = 4.21PLGIVMKK724 pKa = 10.32TPTASRR730 pKa = 11.84ATYY733 pKa = 10.23LRR735 pKa = 11.84WSLFATPAIQSDD747 pKa = 3.26TRR749 pKa = 11.84EE750 pKa = 4.14YY751 pKa = 10.64PFGGYY756 pKa = 9.04SRR758 pKa = 11.84SGTEE762 pKa = 4.06TNHH765 pKa = 6.37HH766 pKa = 6.4VLTALSGGRR775 pKa = 11.84IVVLKK780 pKa = 10.63DD781 pKa = 3.28AEE783 pKa = 4.43AEE785 pKa = 4.14QNPVFLNRR793 pKa = 11.84TSGQPTTNTPQVDD806 pKa = 4.14SQGSTTEE813 pKa = 3.75AFLDD817 pKa = 3.61II818 pKa = 5.09
MM1 pKa = 7.94RR2 pKa = 11.84IAGDD6 pKa = 3.59RR7 pKa = 11.84QTVLLGLPSGKK18 pKa = 10.44GGISVLVGDD27 pKa = 4.92FATNLEE33 pKa = 4.39SVARR37 pKa = 11.84AIFTDD42 pKa = 3.25TNMVVDD48 pKa = 3.85ATIEE52 pKa = 3.82DD53 pKa = 3.73RR54 pKa = 11.84ARR56 pKa = 11.84ARR58 pKa = 11.84NHH60 pKa = 5.73TILRR64 pKa = 11.84EE65 pKa = 3.96GLIDD69 pKa = 3.29SRR71 pKa = 11.84MYY73 pKa = 10.1MSAGRR78 pKa = 11.84TTQVALWAAGPPVAVGVNPTWYY100 pKa = 9.41ILRR103 pKa = 11.84PGKK106 pKa = 10.53VEE108 pKa = 5.32GIMLTQSSSMASLAPLFRR126 pKa = 11.84GVEE129 pKa = 4.07LGPFASAVRR138 pKa = 11.84ADD140 pKa = 3.33VGFYY144 pKa = 11.11SMNVVLGALRR154 pKa = 11.84AAGFNAEE161 pKa = 4.06NGRR164 pKa = 11.84VSFVEE169 pKa = 3.52PLLRR173 pKa = 11.84VQCKK177 pKa = 9.93ILDD180 pKa = 4.03GQDD183 pKa = 3.3RR184 pKa = 11.84GEE186 pKa = 4.13SPYY189 pKa = 10.91SIVGGLTSQLVNPVTQRR206 pKa = 11.84VWGNFFPGGPNQAAWVAADD225 pKa = 3.54TVSRR229 pKa = 11.84VVNIEE234 pKa = 3.59DD235 pKa = 4.31FMRR238 pKa = 11.84EE239 pKa = 3.85AQGEE243 pKa = 4.2NRR245 pKa = 11.84FAAGWGPGFWNKK257 pKa = 9.26MGATGVAVVPIKK269 pKa = 10.24TSGAMLAQQNSAWTLAHH286 pKa = 6.09MEE288 pKa = 4.15YY289 pKa = 9.73PRR291 pKa = 11.84KK292 pKa = 9.76VRR294 pKa = 11.84QVDD297 pKa = 3.69TNVISINAALAAVGAEE313 pKa = 3.66RR314 pKa = 11.84EE315 pKa = 4.29YY316 pKa = 11.22TNTSCTHH323 pKa = 4.78VPGPYY328 pKa = 10.11GKK330 pKa = 10.0VLFVIVDD337 pKa = 3.51QTTDD341 pKa = 2.92RR342 pKa = 11.84AVQVAVEE349 pKa = 4.23GAVGVLLTTPALHH362 pKa = 5.68YY363 pKa = 10.58VIGGIDD369 pKa = 3.4MGIGLMVTRR378 pKa = 11.84EE379 pKa = 3.76VGLAIDD385 pKa = 3.68QAALEE390 pKa = 4.89NIMAMEE396 pKa = 4.43RR397 pKa = 11.84WVEE400 pKa = 4.04FYY402 pKa = 11.31GNEE405 pKa = 4.14EE406 pKa = 4.21DD407 pKa = 3.64WQSATTIVGCAYY419 pKa = 9.21VTFGVEE425 pKa = 4.15MRR427 pKa = 11.84RR428 pKa = 11.84HH429 pKa = 4.81TNRR432 pKa = 11.84VEE434 pKa = 3.95GGYY437 pKa = 9.44YY438 pKa = 8.16WQVGANPPFPLGGLANWTRR457 pKa = 11.84LDD459 pKa = 3.77NLTAAQQAAIYY470 pKa = 10.76ADD472 pKa = 3.14IQNTVEE478 pKa = 4.15APWGGFRR485 pKa = 11.84VRR487 pKa = 11.84NPVPTAAPNAMYY499 pKa = 9.96TRR501 pKa = 11.84FEE503 pKa = 4.06PVLAVGTVAMLWRR516 pKa = 11.84PEE518 pKa = 4.05HH519 pKa = 6.85PMLAPLPDD527 pKa = 4.19RR528 pKa = 11.84PSALADD534 pKa = 2.85ICYY537 pKa = 10.13QRR539 pKa = 11.84GRR541 pKa = 11.84MCAAQEE547 pKa = 4.46DD548 pKa = 4.94IVVGQLGFPEE558 pKa = 4.25DD559 pKa = 4.03MIRR562 pKa = 11.84APVGVGNVGLAQRR575 pKa = 11.84LIRR578 pKa = 11.84KK579 pKa = 6.56MCNVNNQILEE589 pKa = 4.27AKK591 pKa = 9.46HH592 pKa = 6.02ASTVHH597 pKa = 5.22VVLNGYY603 pKa = 8.67WSIGAGVTYY612 pKa = 10.28RR613 pKa = 11.84AIYY616 pKa = 9.01AQSGPGTMRR625 pKa = 11.84KK626 pKa = 9.71RR627 pKa = 11.84IPWAEE632 pKa = 3.87MRR634 pKa = 11.84SLGFDD639 pKa = 3.36ADD641 pKa = 4.05LVKK644 pKa = 10.94NDD646 pKa = 4.2LYY648 pKa = 11.46GIGSGDD654 pKa = 3.46FVAVNATIGALGGIWDD670 pKa = 3.95VVRR673 pKa = 11.84YY674 pKa = 7.15PLPVPSKK681 pKa = 9.12EE682 pKa = 3.71ARR684 pKa = 11.84AAMNTTNCMGGWTRR698 pKa = 11.84AVTRR702 pKa = 11.84VDD704 pKa = 3.05NLGTIRR710 pKa = 11.84EE711 pKa = 4.46GGLEE715 pKa = 4.14PEE717 pKa = 4.21PLGIVMKK724 pKa = 10.32TPTASRR730 pKa = 11.84ATYY733 pKa = 10.23LRR735 pKa = 11.84WSLFATPAIQSDD747 pKa = 3.26TRR749 pKa = 11.84EE750 pKa = 4.14YY751 pKa = 10.64PFGGYY756 pKa = 9.04SRR758 pKa = 11.84SGTEE762 pKa = 4.06TNHH765 pKa = 6.37HH766 pKa = 6.4VLTALSGGRR775 pKa = 11.84IVVLKK780 pKa = 10.63DD781 pKa = 3.28AEE783 pKa = 4.43AEE785 pKa = 4.14QNPVFLNRR793 pKa = 11.84TSGQPTTNTPQVDD806 pKa = 4.14SQGSTTEE813 pKa = 3.75AFLDD817 pKa = 3.61II818 pKa = 5.09
Molecular weight: 88.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A125SJD3|A0A125SJD3_9VIRU Structural protein OS=Piscine myocarditis-like virus OX=1798085 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 10.18GACRR6 pKa = 11.84YY7 pKa = 8.01TMEE10 pKa = 4.24IYY12 pKa = 10.8FNFKK16 pKa = 10.29CSNKK20 pKa = 9.99SLSLMLGFILIMYY33 pKa = 9.13LMSLLQKK40 pKa = 10.78VGAQPLALKK49 pKa = 9.03VTEE52 pKa = 4.78TYY54 pKa = 10.83CPTFLTLKK62 pKa = 10.6VPDD65 pKa = 4.31KK66 pKa = 10.61LISSMKK72 pKa = 9.11TVTGGWIVKK81 pKa = 8.0TAAGRR86 pKa = 11.84EE87 pKa = 4.21ICVHH91 pKa = 5.83GDD93 pKa = 2.2WWEE96 pKa = 3.88AQFNLLGKK104 pKa = 10.41GRR106 pKa = 11.84LEE108 pKa = 4.38RR109 pKa = 11.84ITLHH113 pKa = 4.6GHH115 pKa = 4.97EE116 pKa = 4.61VLVPTKK122 pKa = 10.62FEE124 pKa = 4.05IKK126 pKa = 10.48VNEE129 pKa = 4.19DD130 pKa = 2.92HH131 pKa = 7.77SIVQTNVNCANGIGTWNLFGRR152 pKa = 11.84VDD154 pKa = 3.72CTVVEE159 pKa = 4.41NVVQGHH165 pKa = 6.69DD166 pKa = 3.52NQAAMAAVPPAQPPPPPLQTLWSEE190 pKa = 3.97IVGWGTDD197 pKa = 3.36TLILIGIVLALILIIWKK214 pKa = 9.18GVPCVVRR221 pKa = 11.84LIVKK225 pKa = 9.98RR226 pKa = 11.84KK227 pKa = 9.55KK228 pKa = 10.2KK229 pKa = 10.25NKK231 pKa = 8.38QKK233 pKa = 11.01RR234 pKa = 11.84KK235 pKa = 8.93IKK237 pKa = 10.23KK238 pKa = 9.21KK239 pKa = 10.03KK240 pKa = 9.49KK241 pKa = 9.33KK242 pKa = 9.81KK243 pKa = 9.68KK244 pKa = 9.63KK245 pKa = 9.63KK246 pKa = 9.63KK247 pKa = 9.63KK248 pKa = 9.63KK249 pKa = 9.63KK250 pKa = 9.63KK251 pKa = 9.63KK252 pKa = 9.63KK253 pKa = 9.63KK254 pKa = 9.48KK255 pKa = 9.26KK256 pKa = 9.28KK257 pKa = 9.31KK258 pKa = 9.95KK259 pKa = 8.27KK260 pKa = 7.97TFLIIYY266 pKa = 6.93SQKK269 pKa = 10.53KK270 pKa = 9.03KK271 pKa = 10.46KK272 pKa = 10.43KK273 pKa = 8.48KK274 pKa = 8.29TKK276 pKa = 9.07KK277 pKa = 10.19QNTILHH283 pKa = 6.58LSYY286 pKa = 10.95IKK288 pKa = 10.24IKK290 pKa = 10.83KK291 pKa = 7.53KK292 pKa = 8.44TKK294 pKa = 9.6KK295 pKa = 9.86KK296 pKa = 10.07KK297 pKa = 9.62KK298 pKa = 9.88KK299 pKa = 8.17KK300 pKa = 8.74TKK302 pKa = 10.21
MM1 pKa = 7.58KK2 pKa = 10.18GACRR6 pKa = 11.84YY7 pKa = 8.01TMEE10 pKa = 4.24IYY12 pKa = 10.8FNFKK16 pKa = 10.29CSNKK20 pKa = 9.99SLSLMLGFILIMYY33 pKa = 9.13LMSLLQKK40 pKa = 10.78VGAQPLALKK49 pKa = 9.03VTEE52 pKa = 4.78TYY54 pKa = 10.83CPTFLTLKK62 pKa = 10.6VPDD65 pKa = 4.31KK66 pKa = 10.61LISSMKK72 pKa = 9.11TVTGGWIVKK81 pKa = 8.0TAAGRR86 pKa = 11.84EE87 pKa = 4.21ICVHH91 pKa = 5.83GDD93 pKa = 2.2WWEE96 pKa = 3.88AQFNLLGKK104 pKa = 10.41GRR106 pKa = 11.84LEE108 pKa = 4.38RR109 pKa = 11.84ITLHH113 pKa = 4.6GHH115 pKa = 4.97EE116 pKa = 4.61VLVPTKK122 pKa = 10.62FEE124 pKa = 4.05IKK126 pKa = 10.48VNEE129 pKa = 4.19DD130 pKa = 2.92HH131 pKa = 7.77SIVQTNVNCANGIGTWNLFGRR152 pKa = 11.84VDD154 pKa = 3.72CTVVEE159 pKa = 4.41NVVQGHH165 pKa = 6.69DD166 pKa = 3.52NQAAMAAVPPAQPPPPPLQTLWSEE190 pKa = 3.97IVGWGTDD197 pKa = 3.36TLILIGIVLALILIIWKK214 pKa = 9.18GVPCVVRR221 pKa = 11.84LIVKK225 pKa = 9.98RR226 pKa = 11.84KK227 pKa = 9.55KK228 pKa = 10.2KK229 pKa = 10.25NKK231 pKa = 8.38QKK233 pKa = 11.01RR234 pKa = 11.84KK235 pKa = 8.93IKK237 pKa = 10.23KK238 pKa = 9.21KK239 pKa = 10.03KK240 pKa = 9.49KK241 pKa = 9.33KK242 pKa = 9.81KK243 pKa = 9.68KK244 pKa = 9.63KK245 pKa = 9.63KK246 pKa = 9.63KK247 pKa = 9.63KK248 pKa = 9.63KK249 pKa = 9.63KK250 pKa = 9.63KK251 pKa = 9.63KK252 pKa = 9.63KK253 pKa = 9.63KK254 pKa = 9.48KK255 pKa = 9.26KK256 pKa = 9.28KK257 pKa = 9.31KK258 pKa = 9.95KK259 pKa = 8.27KK260 pKa = 7.97TFLIIYY266 pKa = 6.93SQKK269 pKa = 10.53KK270 pKa = 9.03KK271 pKa = 10.46KK272 pKa = 10.43KK273 pKa = 8.48KK274 pKa = 8.29TKK276 pKa = 9.07KK277 pKa = 10.19QNTILHH283 pKa = 6.58LSYY286 pKa = 10.95IKK288 pKa = 10.24IKK290 pKa = 10.83KK291 pKa = 7.53KK292 pKa = 8.44TKK294 pKa = 9.6KK295 pKa = 9.86KK296 pKa = 10.07KK297 pKa = 9.62KK298 pKa = 9.88KK299 pKa = 8.17KK300 pKa = 8.74TKK302 pKa = 10.21
Molecular weight: 34.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1951 |
302 |
831 |
650.3 |
72.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.381 ± 2.091 | 1.076 ± 0.245 |
4.511 ± 0.778 | 5.331 ± 0.715 |
3.434 ± 0.365 | 8.303 ± 1.07 |
1.23 ± 0.171 | 5.997 ± 0.668 |
7.278 ± 3.326 | 8.303 ± 0.594 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.717 ± 0.331 | 4.664 ± 0.149 |
4.408 ± 0.721 | 3.178 ± 0.45 |
6.509 ± 0.826 | 5.894 ± 1.276 |
6.048 ± 0.785 | 8.662 ± 0.765 |
2.46 ± 0.183 | 2.614 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
