
Sanxia Water Strider Virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
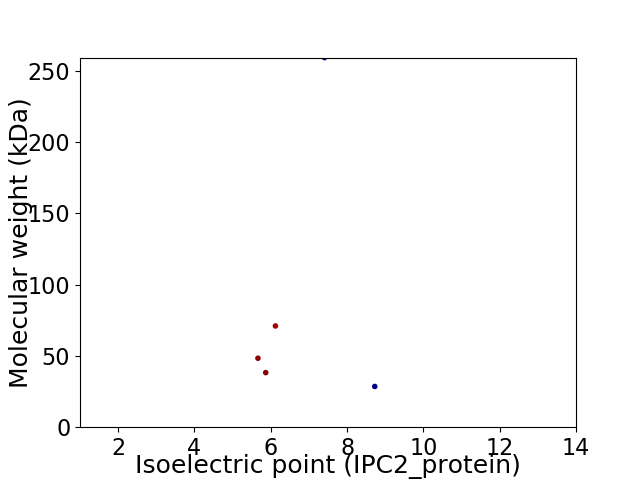
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KTE8|A0A0B5KTE8_9RHAB ORF2 OS=Sanxia Water Strider Virus 5 OX=1608064 GN=ORF2 PE=4 SV=1
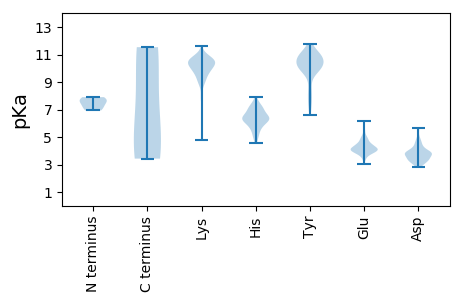
MM1 pKa = 7.21TSSIEE6 pKa = 4.27EE7 pKa = 3.59IWKK10 pKa = 10.31VEE12 pKa = 3.75GLEE15 pKa = 4.12KK16 pKa = 10.82VSAVTVGVTRR26 pKa = 11.84VAWSDD31 pKa = 3.35QYY33 pKa = 11.74FLSTYY38 pKa = 9.9FKK40 pKa = 10.07TIRR43 pKa = 11.84ARR45 pKa = 11.84SSATAARR52 pKa = 11.84TACRR56 pKa = 11.84VFLRR60 pKa = 11.84WYY62 pKa = 10.62VGGCLPEE69 pKa = 4.07NVKK72 pKa = 11.0LIDD75 pKa = 4.63LYY77 pKa = 11.58LSIISKK83 pKa = 9.68MSYY86 pKa = 10.78GEE88 pKa = 4.0NHH90 pKa = 6.53SLFPDD95 pKa = 3.4SGFNGATSVPIGNLGTPGTQDD116 pKa = 3.78LEE118 pKa = 4.52YY119 pKa = 10.76KK120 pKa = 9.85NNHH123 pKa = 4.55VTTLEE128 pKa = 4.24LTSSSTGAPLLWNDD142 pKa = 3.4TSVKK146 pKa = 9.15EE147 pKa = 3.79WHH149 pKa = 6.35NRR151 pKa = 11.84LLEE154 pKa = 3.95YY155 pKa = 10.53DD156 pKa = 3.62ISNSPDD162 pKa = 3.22EE163 pKa = 4.49FSEE166 pKa = 4.2SQRR169 pKa = 11.84NFVNICGFLALTMLRR184 pKa = 11.84GIAKK188 pKa = 9.8DD189 pKa = 2.91ADD191 pKa = 3.81MITRR195 pKa = 11.84SMAKK199 pKa = 8.81TVAANINNLWNLTLSEE215 pKa = 4.55CPPPNMQSIAYY226 pKa = 8.07MMTVMKK232 pKa = 10.41KK233 pKa = 10.46GSPNANLILGQILNSYY249 pKa = 8.82MDD251 pKa = 3.4QSTNPTVVGIFRR263 pKa = 11.84ASCLMSLGMNGMNSLHH279 pKa = 6.4WLMSAARR286 pKa = 11.84VNNIQPIVIARR297 pKa = 11.84CLATNSHH304 pKa = 6.29SKK306 pKa = 9.73TNTAIVDD313 pKa = 3.62LMKK316 pKa = 11.04NQMLAGQTTWEE327 pKa = 3.86WSRR330 pKa = 11.84LFNEE334 pKa = 4.84GAFMDD339 pKa = 4.24LSVRR343 pKa = 11.84EE344 pKa = 4.15DD345 pKa = 3.44PLYY348 pKa = 11.1AGTCFFLSEE357 pKa = 4.09GHH359 pKa = 7.14DD360 pKa = 3.82AQPSNYY366 pKa = 9.58LSLASYY372 pKa = 10.93LKK374 pKa = 11.04GPILDD379 pKa = 3.71SAKK382 pKa = 10.32IKK384 pKa = 10.28ALKK387 pKa = 8.81IAAYY391 pKa = 10.23LRR393 pKa = 11.84EE394 pKa = 4.12QDD396 pKa = 4.51LSTAQTTEE404 pKa = 3.39ASAVANVIIQTPTRR418 pKa = 11.84LDD420 pKa = 3.38VHH422 pKa = 6.19HH423 pKa = 6.6TAIPSLNQGAGDD435 pKa = 3.46MDD437 pKa = 4.08EE438 pKa = 4.27FF439 pKa = 6.3
MM1 pKa = 7.21TSSIEE6 pKa = 4.27EE7 pKa = 3.59IWKK10 pKa = 10.31VEE12 pKa = 3.75GLEE15 pKa = 4.12KK16 pKa = 10.82VSAVTVGVTRR26 pKa = 11.84VAWSDD31 pKa = 3.35QYY33 pKa = 11.74FLSTYY38 pKa = 9.9FKK40 pKa = 10.07TIRR43 pKa = 11.84ARR45 pKa = 11.84SSATAARR52 pKa = 11.84TACRR56 pKa = 11.84VFLRR60 pKa = 11.84WYY62 pKa = 10.62VGGCLPEE69 pKa = 4.07NVKK72 pKa = 11.0LIDD75 pKa = 4.63LYY77 pKa = 11.58LSIISKK83 pKa = 9.68MSYY86 pKa = 10.78GEE88 pKa = 4.0NHH90 pKa = 6.53SLFPDD95 pKa = 3.4SGFNGATSVPIGNLGTPGTQDD116 pKa = 3.78LEE118 pKa = 4.52YY119 pKa = 10.76KK120 pKa = 9.85NNHH123 pKa = 4.55VTTLEE128 pKa = 4.24LTSSSTGAPLLWNDD142 pKa = 3.4TSVKK146 pKa = 9.15EE147 pKa = 3.79WHH149 pKa = 6.35NRR151 pKa = 11.84LLEE154 pKa = 3.95YY155 pKa = 10.53DD156 pKa = 3.62ISNSPDD162 pKa = 3.22EE163 pKa = 4.49FSEE166 pKa = 4.2SQRR169 pKa = 11.84NFVNICGFLALTMLRR184 pKa = 11.84GIAKK188 pKa = 9.8DD189 pKa = 2.91ADD191 pKa = 3.81MITRR195 pKa = 11.84SMAKK199 pKa = 8.81TVAANINNLWNLTLSEE215 pKa = 4.55CPPPNMQSIAYY226 pKa = 8.07MMTVMKK232 pKa = 10.41KK233 pKa = 10.46GSPNANLILGQILNSYY249 pKa = 8.82MDD251 pKa = 3.4QSTNPTVVGIFRR263 pKa = 11.84ASCLMSLGMNGMNSLHH279 pKa = 6.4WLMSAARR286 pKa = 11.84VNNIQPIVIARR297 pKa = 11.84CLATNSHH304 pKa = 6.29SKK306 pKa = 9.73TNTAIVDD313 pKa = 3.62LMKK316 pKa = 11.04NQMLAGQTTWEE327 pKa = 3.86WSRR330 pKa = 11.84LFNEE334 pKa = 4.84GAFMDD339 pKa = 4.24LSVRR343 pKa = 11.84EE344 pKa = 4.15DD345 pKa = 3.44PLYY348 pKa = 11.1AGTCFFLSEE357 pKa = 4.09GHH359 pKa = 7.14DD360 pKa = 3.82AQPSNYY366 pKa = 9.58LSLASYY372 pKa = 10.93LKK374 pKa = 11.04GPILDD379 pKa = 3.71SAKK382 pKa = 10.32IKK384 pKa = 10.28ALKK387 pKa = 8.81IAAYY391 pKa = 10.23LRR393 pKa = 11.84EE394 pKa = 4.12QDD396 pKa = 4.51LSTAQTTEE404 pKa = 3.39ASAVANVIIQTPTRR418 pKa = 11.84LDD420 pKa = 3.38VHH422 pKa = 6.19HH423 pKa = 6.6TAIPSLNQGAGDD435 pKa = 3.46MDD437 pKa = 4.08EE438 pKa = 4.27FF439 pKa = 6.3
Molecular weight: 48.36 kDa
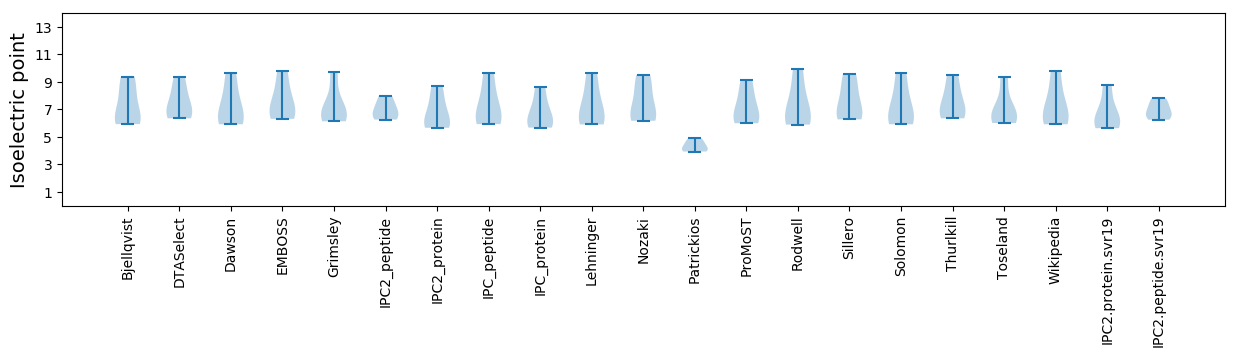
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KXE2|A0A0B5KXE2_9RHAB ORF3 OS=Sanxia Water Strider Virus 5 OX=1608064 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.9DD2 pKa = 5.28LSKK5 pKa = 10.64IHH7 pKa = 6.04NQVSYY12 pKa = 10.02TKK14 pKa = 10.63VKK16 pKa = 10.4GRR18 pKa = 11.84LLMSFEE24 pKa = 4.23LTGLQSKK31 pKa = 9.12SWSVYY36 pKa = 10.15KK37 pKa = 8.77YY38 pKa = 9.97VCGLVILEE46 pKa = 4.78LFYY49 pKa = 11.62NMEE52 pKa = 3.84VSEE55 pKa = 4.56IFLDD59 pKa = 4.05SVISNLYY66 pKa = 9.74QRR68 pKa = 11.84TKK70 pKa = 10.25EE71 pKa = 4.02VPVTYY76 pKa = 10.0RR77 pKa = 11.84PSPSDD82 pKa = 3.16SLLRR86 pKa = 11.84GVVEE90 pKa = 4.79LKK92 pKa = 10.15WFTASACFPTTHH104 pKa = 7.01KK105 pKa = 10.21KK106 pKa = 10.75FNMNKK111 pKa = 9.56SRR113 pKa = 11.84TFPLISLGSQQILISYY129 pKa = 7.89DD130 pKa = 3.24FRR132 pKa = 11.84SSIEE136 pKa = 4.0PYY138 pKa = 10.63DD139 pKa = 3.94PEE141 pKa = 4.1NLIALLEE148 pKa = 4.4TGHH151 pKa = 7.6INIEE155 pKa = 3.93KK156 pKa = 10.33HH157 pKa = 6.46SADD160 pKa = 3.62LHH162 pKa = 5.98CSKK165 pKa = 10.54ARR167 pKa = 11.84KK168 pKa = 9.11QIHH171 pKa = 4.97TVRR174 pKa = 11.84KK175 pKa = 4.81TTHH178 pKa = 4.87VLNYY182 pKa = 9.92TEE184 pKa = 5.17LLNLVQMDD192 pKa = 3.52VDD194 pKa = 4.32LVKK197 pKa = 10.65AANTRR202 pKa = 11.84KK203 pKa = 10.04AEE205 pKa = 4.17KK206 pKa = 10.45NFLPTDD212 pKa = 3.79PVHH215 pKa = 6.72KK216 pKa = 9.89KK217 pKa = 10.71VEE219 pKa = 4.28VGHH222 pKa = 5.99QMLGRR227 pKa = 11.84QAPNYY232 pKa = 7.0EE233 pKa = 3.74VKK235 pKa = 9.44MRR237 pKa = 11.84KK238 pKa = 9.17GATLSKK244 pKa = 10.51SNCIVSS250 pKa = 3.43
MM1 pKa = 7.9DD2 pKa = 5.28LSKK5 pKa = 10.64IHH7 pKa = 6.04NQVSYY12 pKa = 10.02TKK14 pKa = 10.63VKK16 pKa = 10.4GRR18 pKa = 11.84LLMSFEE24 pKa = 4.23LTGLQSKK31 pKa = 9.12SWSVYY36 pKa = 10.15KK37 pKa = 8.77YY38 pKa = 9.97VCGLVILEE46 pKa = 4.78LFYY49 pKa = 11.62NMEE52 pKa = 3.84VSEE55 pKa = 4.56IFLDD59 pKa = 4.05SVISNLYY66 pKa = 9.74QRR68 pKa = 11.84TKK70 pKa = 10.25EE71 pKa = 4.02VPVTYY76 pKa = 10.0RR77 pKa = 11.84PSPSDD82 pKa = 3.16SLLRR86 pKa = 11.84GVVEE90 pKa = 4.79LKK92 pKa = 10.15WFTASACFPTTHH104 pKa = 7.01KK105 pKa = 10.21KK106 pKa = 10.75FNMNKK111 pKa = 9.56SRR113 pKa = 11.84TFPLISLGSQQILISYY129 pKa = 7.89DD130 pKa = 3.24FRR132 pKa = 11.84SSIEE136 pKa = 4.0PYY138 pKa = 10.63DD139 pKa = 3.94PEE141 pKa = 4.1NLIALLEE148 pKa = 4.4TGHH151 pKa = 7.6INIEE155 pKa = 3.93KK156 pKa = 10.33HH157 pKa = 6.46SADD160 pKa = 3.62LHH162 pKa = 5.98CSKK165 pKa = 10.54ARR167 pKa = 11.84KK168 pKa = 9.11QIHH171 pKa = 4.97TVRR174 pKa = 11.84KK175 pKa = 4.81TTHH178 pKa = 4.87VLNYY182 pKa = 9.92TEE184 pKa = 5.17LLNLVQMDD192 pKa = 3.52VDD194 pKa = 4.32LVKK197 pKa = 10.65AANTRR202 pKa = 11.84KK203 pKa = 10.04AEE205 pKa = 4.17KK206 pKa = 10.45NFLPTDD212 pKa = 3.79PVHH215 pKa = 6.72KK216 pKa = 9.89KK217 pKa = 10.71VEE219 pKa = 4.28VGHH222 pKa = 5.99QMLGRR227 pKa = 11.84QAPNYY232 pKa = 7.0EE233 pKa = 3.74VKK235 pKa = 9.44MRR237 pKa = 11.84KK238 pKa = 9.17GATLSKK244 pKa = 10.51SNCIVSS250 pKa = 3.43
Molecular weight: 28.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3917 |
250 |
2264 |
783.4 |
89.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.493 ± 0.737 | 1.557 ± 0.153 |
4.468 ± 0.181 | 5.438 ± 0.36 |
4.723 ± 0.532 | 4.468 ± 0.345 |
2.885 ± 0.259 | 7.123 ± 0.647 |
5.948 ± 0.802 | 12.076 ± 0.75 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.604 ± 0.296 | 4.953 ± 0.475 |
5.234 ± 0.416 | 3.778 ± 0.339 |
4.187 ± 0.35 | 10.008 ± 0.654 |
6.382 ± 0.512 | 4.621 ± 0.616 |
1.583 ± 0.139 | 3.472 ± 0.21 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
