
Rasamsonia emersonii CBS 393.64
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Trichocomaceae; Rasamsonia; Rasamsonia emersonii
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9842 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
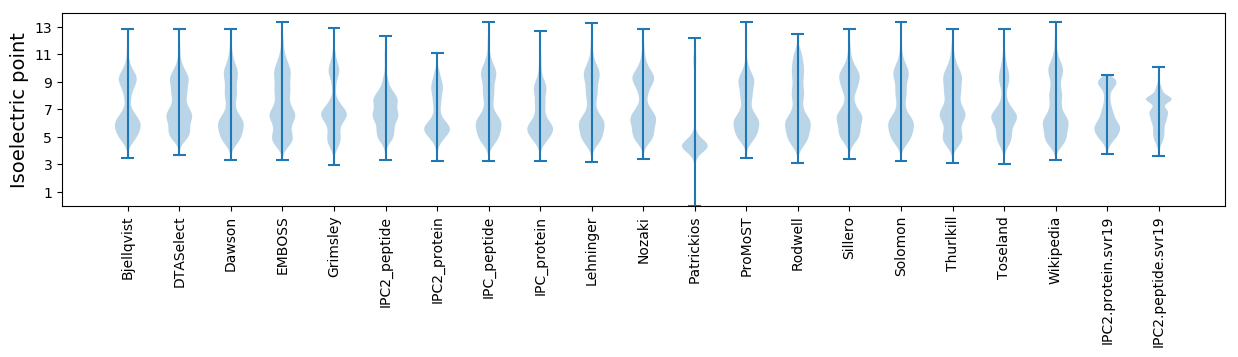
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0F4Z3Q1|A0A0F4Z3Q1_TALEM WD repeat protein OS=Rasamsonia emersonii CBS 393.64 OX=1408163 GN=T310_0832 PE=4 SV=1
MM1 pKa = 7.55PGIYY5 pKa = 9.66TLVTLSTLLTSINVLAAPAVPATDD29 pKa = 4.3APAPTPPPKK38 pKa = 10.42LEE40 pKa = 4.07EE41 pKa = 4.1RR42 pKa = 11.84ATTCTFSGSDD52 pKa = 3.63GASSASKK59 pKa = 10.75SKK61 pKa = 9.53TSCSTIVLSDD71 pKa = 3.44VAVPSGTTLDD81 pKa = 3.9LTNLNDD87 pKa = 3.64GTHH90 pKa = 6.34VIFEE94 pKa = 4.49GEE96 pKa = 4.12TTFGYY101 pKa = 10.01EE102 pKa = 3.84EE103 pKa = 4.09WSGPLVSVSGTDD115 pKa = 3.13ITVTAASGATLNGDD129 pKa = 4.09GSRR132 pKa = 11.84WWDD135 pKa = 3.42GEE137 pKa = 3.87GSNGGKK143 pKa = 8.26TKK145 pKa = 10.59PKK147 pKa = 10.16FFYY150 pKa = 10.65AHH152 pKa = 6.24NLKK155 pKa = 10.56LSSISGLYY163 pKa = 8.66IQNSPVQVFSISGSEE178 pKa = 4.32DD179 pKa = 3.04LTLSDD184 pKa = 3.91ITIDD188 pKa = 5.54DD189 pKa = 3.95SAGDD193 pKa = 3.59NAGAANTDD201 pKa = 3.3GFDD204 pKa = 3.34VGEE207 pKa = 4.16SSGIVITGATVYY219 pKa = 10.94NQDD222 pKa = 3.4DD223 pKa = 4.15CLAVNSGTNITFSGGLCSGGHH244 pKa = 5.81GLSIGSVGGRR254 pKa = 11.84SDD256 pKa = 3.49NTVSDD261 pKa = 3.72VTIEE265 pKa = 3.95SSQVKK270 pKa = 10.05NSQNGVRR277 pKa = 11.84IKK279 pKa = 9.73TVYY282 pKa = 10.35DD283 pKa = 3.27ATGSVSGVTYY293 pKa = 10.99KK294 pKa = 10.92DD295 pKa = 3.0ITLSGITDD303 pKa = 3.35YY304 pKa = 11.65GIVVRR309 pKa = 11.84QDD311 pKa = 3.47YY312 pKa = 11.2EE313 pKa = 4.21NGSPTGTPTNGVPISDD329 pKa = 3.64FTLDD333 pKa = 3.63NVQGTVDD340 pKa = 4.09SDD342 pKa = 3.41ATDD345 pKa = 3.17IFILCGAGSCSDD357 pKa = 3.6WTWTDD362 pKa = 3.28VNVTGGKK369 pKa = 10.1VSDD372 pKa = 4.03DD373 pKa = 4.12CEE375 pKa = 4.19NVPSGISCSAA385 pKa = 3.3
MM1 pKa = 7.55PGIYY5 pKa = 9.66TLVTLSTLLTSINVLAAPAVPATDD29 pKa = 4.3APAPTPPPKK38 pKa = 10.42LEE40 pKa = 4.07EE41 pKa = 4.1RR42 pKa = 11.84ATTCTFSGSDD52 pKa = 3.63GASSASKK59 pKa = 10.75SKK61 pKa = 9.53TSCSTIVLSDD71 pKa = 3.44VAVPSGTTLDD81 pKa = 3.9LTNLNDD87 pKa = 3.64GTHH90 pKa = 6.34VIFEE94 pKa = 4.49GEE96 pKa = 4.12TTFGYY101 pKa = 10.01EE102 pKa = 3.84EE103 pKa = 4.09WSGPLVSVSGTDD115 pKa = 3.13ITVTAASGATLNGDD129 pKa = 4.09GSRR132 pKa = 11.84WWDD135 pKa = 3.42GEE137 pKa = 3.87GSNGGKK143 pKa = 8.26TKK145 pKa = 10.59PKK147 pKa = 10.16FFYY150 pKa = 10.65AHH152 pKa = 6.24NLKK155 pKa = 10.56LSSISGLYY163 pKa = 8.66IQNSPVQVFSISGSEE178 pKa = 4.32DD179 pKa = 3.04LTLSDD184 pKa = 3.91ITIDD188 pKa = 5.54DD189 pKa = 3.95SAGDD193 pKa = 3.59NAGAANTDD201 pKa = 3.3GFDD204 pKa = 3.34VGEE207 pKa = 4.16SSGIVITGATVYY219 pKa = 10.94NQDD222 pKa = 3.4DD223 pKa = 4.15CLAVNSGTNITFSGGLCSGGHH244 pKa = 5.81GLSIGSVGGRR254 pKa = 11.84SDD256 pKa = 3.49NTVSDD261 pKa = 3.72VTIEE265 pKa = 3.95SSQVKK270 pKa = 10.05NSQNGVRR277 pKa = 11.84IKK279 pKa = 9.73TVYY282 pKa = 10.35DD283 pKa = 3.27ATGSVSGVTYY293 pKa = 10.99KK294 pKa = 10.92DD295 pKa = 3.0ITLSGITDD303 pKa = 3.35YY304 pKa = 11.65GIVVRR309 pKa = 11.84QDD311 pKa = 3.47YY312 pKa = 11.2EE313 pKa = 4.21NGSPTGTPTNGVPISDD329 pKa = 3.64FTLDD333 pKa = 3.63NVQGTVDD340 pKa = 4.09SDD342 pKa = 3.41ATDD345 pKa = 3.17IFILCGAGSCSDD357 pKa = 3.6WTWTDD362 pKa = 3.28VNVTGGKK369 pKa = 10.1VSDD372 pKa = 4.03DD373 pKa = 4.12CEE375 pKa = 4.19NVPSGISCSAA385 pKa = 3.3
Molecular weight: 39.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F4YHL6|A0A0F4YHL6_TALEM Protein kinase (Fragment) OS=Rasamsonia emersonii CBS 393.64 OX=1408163 GN=T310_8391 PE=4 SV=1
QQ1 pKa = 7.11GLVSRR6 pKa = 11.84LKK8 pKa = 10.62CPRR11 pKa = 11.84PRR13 pKa = 11.84PRR15 pKa = 11.84PRR17 pKa = 11.84LKK19 pKa = 10.39RR20 pKa = 11.84APTRR24 pKa = 11.84LPTRR28 pKa = 11.84HH29 pKa = 5.94SPRR32 pKa = 11.84SRR34 pKa = 11.84MVEE37 pKa = 3.78LPSRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84LLRR46 pKa = 11.84PRR48 pKa = 11.84ARR50 pKa = 11.84KK51 pKa = 9.64SLGSLSPTKK60 pKa = 10.37RR61 pKa = 11.84KK62 pKa = 9.7LLARR66 pKa = 11.84TGRR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84GRR73 pKa = 11.84LLAAPNLNRR82 pKa = 11.84TATRR86 pKa = 11.84RR87 pKa = 11.84WPAA90 pKa = 2.86
QQ1 pKa = 7.11GLVSRR6 pKa = 11.84LKK8 pKa = 10.62CPRR11 pKa = 11.84PRR13 pKa = 11.84PRR15 pKa = 11.84PRR17 pKa = 11.84LKK19 pKa = 10.39RR20 pKa = 11.84APTRR24 pKa = 11.84LPTRR28 pKa = 11.84HH29 pKa = 5.94SPRR32 pKa = 11.84SRR34 pKa = 11.84MVEE37 pKa = 3.78LPSRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84LLRR46 pKa = 11.84PRR48 pKa = 11.84ARR50 pKa = 11.84KK51 pKa = 9.64SLGSLSPTKK60 pKa = 10.37RR61 pKa = 11.84KK62 pKa = 9.7LLARR66 pKa = 11.84TGRR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84GRR73 pKa = 11.84LLAAPNLNRR82 pKa = 11.84TATRR86 pKa = 11.84RR87 pKa = 11.84WPAA90 pKa = 2.86
Molecular weight: 10.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4794008 |
73 |
7865 |
487.1 |
53.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.564 ± 0.021 | 1.255 ± 0.009 |
5.662 ± 0.017 | 6.324 ± 0.027 |
3.589 ± 0.016 | 6.649 ± 0.018 |
2.365 ± 0.01 | 4.743 ± 0.016 |
4.701 ± 0.021 | 8.92 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.961 ± 0.008 | 3.647 ± 0.013 |
6.28 ± 0.027 | 4.138 ± 0.019 |
6.604 ± 0.023 | 8.557 ± 0.03 |
5.782 ± 0.016 | 6.109 ± 0.018 |
1.403 ± 0.009 | 2.745 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
