
Hubei picorna-like virus 67
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
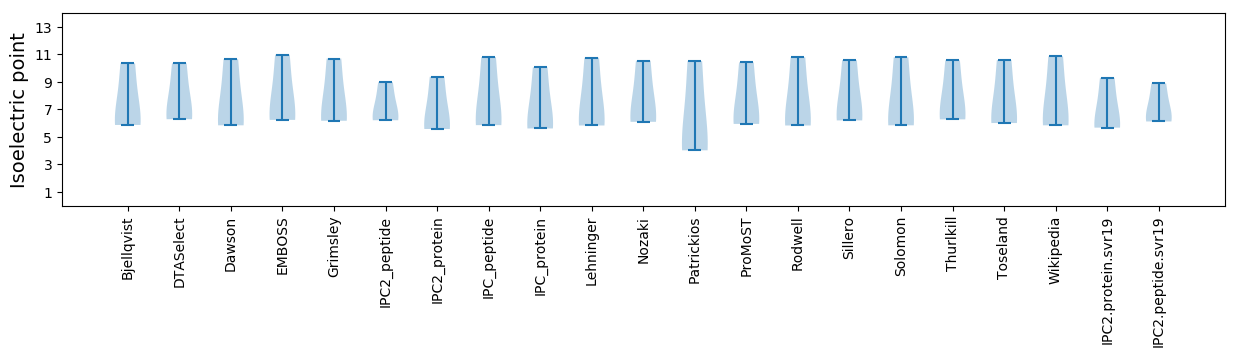
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
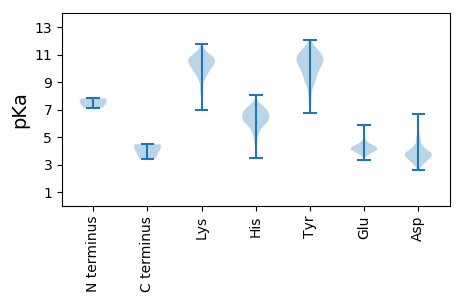
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KM14|A0A1L3KM14_9VIRU Uncharacterized protein OS=Hubei picorna-like virus 67 OX=1923150 PE=4 SV=1
MM1 pKa = 7.82EE2 pKa = 5.31EE3 pKa = 4.43SQDD6 pKa = 3.76QSWGVPVFQPAGLTEE21 pKa = 4.32LSLRR25 pKa = 11.84QISLPTFNEE34 pKa = 4.02DD35 pKa = 3.85QIPPQSFAQEE45 pKa = 3.75RR46 pKa = 11.84RR47 pKa = 11.84EE48 pKa = 3.92LVAQRR53 pKa = 11.84VQLDD57 pKa = 3.63PSLAWAHH64 pKa = 4.97THH66 pKa = 6.71LVAAYY71 pKa = 9.64HH72 pKa = 6.94RR73 pKa = 11.84DD74 pKa = 3.34FNLISNQALMVRR86 pKa = 11.84DD87 pKa = 4.35LCQEE91 pKa = 4.0TVEE94 pKa = 5.7ILAEE98 pKa = 3.84WEE100 pKa = 3.93TRR102 pKa = 11.84MSHH105 pKa = 5.27FRR107 pKa = 11.84KK108 pKa = 9.46IQEE111 pKa = 3.99SQKK114 pKa = 10.84RR115 pKa = 11.84SIAPLSRR122 pKa = 11.84MGLQIAQNMSALICRR137 pKa = 11.84LRR139 pKa = 11.84AKK141 pKa = 9.89QLPPGIQEE149 pKa = 4.16SKK151 pKa = 11.01VLLRR155 pKa = 11.84SLNILLRR162 pKa = 11.84SYY164 pKa = 11.04HH165 pKa = 6.95RR166 pKa = 11.84ILTKK170 pKa = 9.7MCLEE174 pKa = 4.44VSRR177 pKa = 11.84HH178 pKa = 5.35PKK180 pKa = 9.97VEE182 pKa = 3.67IDD184 pKa = 3.51SRR186 pKa = 11.84EE187 pKa = 3.82QALLSLEE194 pKa = 4.31LAHH197 pKa = 6.78CLVILAGEE205 pKa = 4.33SEE207 pKa = 4.56AIEE210 pKa = 4.42YY211 pKa = 10.04YY212 pKa = 10.38HH213 pKa = 6.93LEE215 pKa = 4.03KK216 pKa = 10.28MPEE219 pKa = 3.98VLEE222 pKa = 4.16VEE224 pKa = 4.38PVHH227 pKa = 6.93LNRR230 pKa = 11.84KK231 pKa = 8.77KK232 pKa = 10.65VLAGALQYY240 pKa = 10.68YY241 pKa = 10.08RR242 pKa = 11.84SLLLGQDD249 pKa = 3.71FQSVHH254 pKa = 6.33LSLEE258 pKa = 4.27LEE260 pKa = 4.33SLSMLPKK267 pKa = 9.97IEE269 pKa = 4.36SKK271 pKa = 11.44DD272 pKa = 3.63NITLIWRR279 pKa = 11.84ALRR282 pKa = 11.84HH283 pKa = 5.78NKK285 pKa = 8.35WATLTSSLQEE295 pKa = 4.15FLMPPHH301 pKa = 6.39GPEE304 pKa = 4.0PRR306 pKa = 11.84SDD308 pKa = 3.25RR309 pKa = 11.84YY310 pKa = 10.45LFKK313 pKa = 10.62ISEE316 pKa = 4.18FFPFKK321 pKa = 10.75FFDD324 pKa = 4.03LSQGMLFHH332 pKa = 6.73VVRR335 pKa = 11.84NVQSNHH341 pKa = 3.47VHH343 pKa = 6.15AGEE346 pKa = 3.78IAKK349 pKa = 10.18YY350 pKa = 9.87FSKK353 pKa = 10.74LLKK356 pKa = 10.6EE357 pKa = 4.34EE358 pKa = 4.34IEE360 pKa = 4.06LDD362 pKa = 3.74VNVFEE367 pKa = 4.7TLFGFIIQDD376 pKa = 3.64LQHH379 pKa = 6.72FDD381 pKa = 2.77TWYY384 pKa = 8.59VTIDD388 pKa = 3.52FPRR391 pKa = 11.84QVFDD395 pKa = 4.59LDD397 pKa = 3.77FDD399 pKa = 4.72VVDD402 pKa = 3.57SWRR405 pKa = 11.84TSTLEE410 pKa = 3.78QRR412 pKa = 11.84RR413 pKa = 11.84PDD415 pKa = 4.17DD416 pKa = 4.2GDD418 pKa = 4.68EE419 pKa = 3.94VDD421 pKa = 3.88HH422 pKa = 6.73WFTSRR427 pKa = 11.84IQSDD431 pKa = 3.2MDD433 pKa = 3.45FEE435 pKa = 4.49RR436 pKa = 11.84VMEE439 pKa = 4.36RR440 pKa = 11.84FHH442 pKa = 8.03NPFMLLEE449 pKa = 4.1PQFINLQINQQLINLSYY466 pKa = 9.05TLCSRR471 pKa = 11.84CQVRR475 pKa = 11.84FLNRR479 pKa = 11.84PGAPPNSEE487 pKa = 3.85DD488 pKa = 4.33ALYY491 pKa = 10.72CDD493 pKa = 4.04RR494 pKa = 11.84CRR496 pKa = 11.84NPSFRR501 pKa = 4.26
MM1 pKa = 7.82EE2 pKa = 5.31EE3 pKa = 4.43SQDD6 pKa = 3.76QSWGVPVFQPAGLTEE21 pKa = 4.32LSLRR25 pKa = 11.84QISLPTFNEE34 pKa = 4.02DD35 pKa = 3.85QIPPQSFAQEE45 pKa = 3.75RR46 pKa = 11.84RR47 pKa = 11.84EE48 pKa = 3.92LVAQRR53 pKa = 11.84VQLDD57 pKa = 3.63PSLAWAHH64 pKa = 4.97THH66 pKa = 6.71LVAAYY71 pKa = 9.64HH72 pKa = 6.94RR73 pKa = 11.84DD74 pKa = 3.34FNLISNQALMVRR86 pKa = 11.84DD87 pKa = 4.35LCQEE91 pKa = 4.0TVEE94 pKa = 5.7ILAEE98 pKa = 3.84WEE100 pKa = 3.93TRR102 pKa = 11.84MSHH105 pKa = 5.27FRR107 pKa = 11.84KK108 pKa = 9.46IQEE111 pKa = 3.99SQKK114 pKa = 10.84RR115 pKa = 11.84SIAPLSRR122 pKa = 11.84MGLQIAQNMSALICRR137 pKa = 11.84LRR139 pKa = 11.84AKK141 pKa = 9.89QLPPGIQEE149 pKa = 4.16SKK151 pKa = 11.01VLLRR155 pKa = 11.84SLNILLRR162 pKa = 11.84SYY164 pKa = 11.04HH165 pKa = 6.95RR166 pKa = 11.84ILTKK170 pKa = 9.7MCLEE174 pKa = 4.44VSRR177 pKa = 11.84HH178 pKa = 5.35PKK180 pKa = 9.97VEE182 pKa = 3.67IDD184 pKa = 3.51SRR186 pKa = 11.84EE187 pKa = 3.82QALLSLEE194 pKa = 4.31LAHH197 pKa = 6.78CLVILAGEE205 pKa = 4.33SEE207 pKa = 4.56AIEE210 pKa = 4.42YY211 pKa = 10.04YY212 pKa = 10.38HH213 pKa = 6.93LEE215 pKa = 4.03KK216 pKa = 10.28MPEE219 pKa = 3.98VLEE222 pKa = 4.16VEE224 pKa = 4.38PVHH227 pKa = 6.93LNRR230 pKa = 11.84KK231 pKa = 8.77KK232 pKa = 10.65VLAGALQYY240 pKa = 10.68YY241 pKa = 10.08RR242 pKa = 11.84SLLLGQDD249 pKa = 3.71FQSVHH254 pKa = 6.33LSLEE258 pKa = 4.27LEE260 pKa = 4.33SLSMLPKK267 pKa = 9.97IEE269 pKa = 4.36SKK271 pKa = 11.44DD272 pKa = 3.63NITLIWRR279 pKa = 11.84ALRR282 pKa = 11.84HH283 pKa = 5.78NKK285 pKa = 8.35WATLTSSLQEE295 pKa = 4.15FLMPPHH301 pKa = 6.39GPEE304 pKa = 4.0PRR306 pKa = 11.84SDD308 pKa = 3.25RR309 pKa = 11.84YY310 pKa = 10.45LFKK313 pKa = 10.62ISEE316 pKa = 4.18FFPFKK321 pKa = 10.75FFDD324 pKa = 4.03LSQGMLFHH332 pKa = 6.73VVRR335 pKa = 11.84NVQSNHH341 pKa = 3.47VHH343 pKa = 6.15AGEE346 pKa = 3.78IAKK349 pKa = 10.18YY350 pKa = 9.87FSKK353 pKa = 10.74LLKK356 pKa = 10.6EE357 pKa = 4.34EE358 pKa = 4.34IEE360 pKa = 4.06LDD362 pKa = 3.74VNVFEE367 pKa = 4.7TLFGFIIQDD376 pKa = 3.64LQHH379 pKa = 6.72FDD381 pKa = 2.77TWYY384 pKa = 8.59VTIDD388 pKa = 3.52FPRR391 pKa = 11.84QVFDD395 pKa = 4.59LDD397 pKa = 3.77FDD399 pKa = 4.72VVDD402 pKa = 3.57SWRR405 pKa = 11.84TSTLEE410 pKa = 3.78QRR412 pKa = 11.84RR413 pKa = 11.84PDD415 pKa = 4.17DD416 pKa = 4.2GDD418 pKa = 4.68EE419 pKa = 3.94VDD421 pKa = 3.88HH422 pKa = 6.73WFTSRR427 pKa = 11.84IQSDD431 pKa = 3.2MDD433 pKa = 3.45FEE435 pKa = 4.49RR436 pKa = 11.84VMEE439 pKa = 4.36RR440 pKa = 11.84FHH442 pKa = 8.03NPFMLLEE449 pKa = 4.1PQFINLQINQQLINLSYY466 pKa = 9.05TLCSRR471 pKa = 11.84CQVRR475 pKa = 11.84FLNRR479 pKa = 11.84PGAPPNSEE487 pKa = 3.85DD488 pKa = 4.33ALYY491 pKa = 10.72CDD493 pKa = 4.04RR494 pKa = 11.84CRR496 pKa = 11.84NPSFRR501 pKa = 4.26
Molecular weight: 58.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KM14|A0A1L3KM14_9VIRU Uncharacterized protein OS=Hubei picorna-like virus 67 OX=1923150 PE=4 SV=1
MM1 pKa = 7.56ALFAAEE7 pKa = 4.67EE8 pKa = 4.09ILTTGLRR15 pKa = 11.84PHH17 pKa = 7.25KK18 pKa = 10.16IINNVVRR25 pKa = 11.84PKK27 pKa = 10.23EE28 pKa = 4.13RR29 pKa = 11.84PPPPPNGRR37 pKa = 11.84KK38 pKa = 9.44SGSVLGSAGFSTSWADD54 pKa = 3.57RR55 pKa = 11.84AQFEE59 pKa = 4.78TDD61 pKa = 3.52FVTDD65 pKa = 3.0VQRR68 pKa = 11.84RR69 pKa = 11.84SNTSTILRR77 pKa = 11.84SRR79 pKa = 11.84EE80 pKa = 3.95KK81 pKa = 9.42GTSSATSSIRR91 pKa = 11.84SVSGMGSYY99 pKa = 10.76ASSRR103 pKa = 11.84SLPSRR108 pKa = 11.84FQPDD112 pKa = 3.95FKK114 pKa = 11.18SGFDD118 pKa = 3.5GARR121 pKa = 11.84SVSRR125 pKa = 11.84NSRR128 pKa = 11.84NSSRR132 pKa = 11.84MGDD135 pKa = 3.71SNVSLQEE142 pKa = 3.8NSGVAEE148 pKa = 5.39AINSSIVTHH157 pKa = 6.71GIADD161 pKa = 3.78SSEE164 pKa = 4.27HH165 pKa = 5.93VSADD169 pKa = 3.56LPSPGQAIATGDD181 pKa = 3.55TGVEE185 pKa = 4.51SIAEE189 pKa = 4.17EE190 pKa = 4.4PEE192 pKa = 3.79HH193 pKa = 7.03LITFISSYY201 pKa = 6.78THH203 pKa = 6.32EE204 pKa = 4.56NVPGSEE210 pKa = 4.29SSSEE214 pKa = 4.04SGDD217 pKa = 3.61RR218 pKa = 11.84QSRR221 pKa = 11.84ASTPVSRR228 pKa = 11.84ARR230 pKa = 11.84TLLSNISGRR239 pKa = 11.84IGGNRR244 pKa = 11.84VLPSGEE250 pKa = 4.01NARR253 pKa = 11.84SAGSRR258 pKa = 11.84TGASKK263 pKa = 10.69SKK265 pKa = 9.5EE266 pKa = 3.69GSRR269 pKa = 11.84RR270 pKa = 11.84GPSVLSEE277 pKa = 4.14FAAGAGFSVGPLVSGIGEE295 pKa = 4.61SINAAKK301 pKa = 10.2DD302 pKa = 3.34RR303 pKa = 11.84KK304 pKa = 9.43QRR306 pKa = 11.84QHH308 pKa = 5.98NLNMARR314 pKa = 11.84TSAQQMGYY322 pKa = 9.7PNLLATRR329 pKa = 11.84IPHH332 pKa = 6.09APTWAGTSFRR342 pKa = 4.48
MM1 pKa = 7.56ALFAAEE7 pKa = 4.67EE8 pKa = 4.09ILTTGLRR15 pKa = 11.84PHH17 pKa = 7.25KK18 pKa = 10.16IINNVVRR25 pKa = 11.84PKK27 pKa = 10.23EE28 pKa = 4.13RR29 pKa = 11.84PPPPPNGRR37 pKa = 11.84KK38 pKa = 9.44SGSVLGSAGFSTSWADD54 pKa = 3.57RR55 pKa = 11.84AQFEE59 pKa = 4.78TDD61 pKa = 3.52FVTDD65 pKa = 3.0VQRR68 pKa = 11.84RR69 pKa = 11.84SNTSTILRR77 pKa = 11.84SRR79 pKa = 11.84EE80 pKa = 3.95KK81 pKa = 9.42GTSSATSSIRR91 pKa = 11.84SVSGMGSYY99 pKa = 10.76ASSRR103 pKa = 11.84SLPSRR108 pKa = 11.84FQPDD112 pKa = 3.95FKK114 pKa = 11.18SGFDD118 pKa = 3.5GARR121 pKa = 11.84SVSRR125 pKa = 11.84NSRR128 pKa = 11.84NSSRR132 pKa = 11.84MGDD135 pKa = 3.71SNVSLQEE142 pKa = 3.8NSGVAEE148 pKa = 5.39AINSSIVTHH157 pKa = 6.71GIADD161 pKa = 3.78SSEE164 pKa = 4.27HH165 pKa = 5.93VSADD169 pKa = 3.56LPSPGQAIATGDD181 pKa = 3.55TGVEE185 pKa = 4.51SIAEE189 pKa = 4.17EE190 pKa = 4.4PEE192 pKa = 3.79HH193 pKa = 7.03LITFISSYY201 pKa = 6.78THH203 pKa = 6.32EE204 pKa = 4.56NVPGSEE210 pKa = 4.29SSSEE214 pKa = 4.04SGDD217 pKa = 3.61RR218 pKa = 11.84QSRR221 pKa = 11.84ASTPVSRR228 pKa = 11.84ARR230 pKa = 11.84TLLSNISGRR239 pKa = 11.84IGGNRR244 pKa = 11.84VLPSGEE250 pKa = 4.01NARR253 pKa = 11.84SAGSRR258 pKa = 11.84TGASKK263 pKa = 10.69SKK265 pKa = 9.5EE266 pKa = 3.69GSRR269 pKa = 11.84RR270 pKa = 11.84GPSVLSEE277 pKa = 4.14FAAGAGFSVGPLVSGIGEE295 pKa = 4.61SINAAKK301 pKa = 10.2DD302 pKa = 3.34RR303 pKa = 11.84KK304 pKa = 9.43QRR306 pKa = 11.84QHH308 pKa = 5.98NLNMARR314 pKa = 11.84TSAQQMGYY322 pKa = 9.7PNLLATRR329 pKa = 11.84IPHH332 pKa = 6.09APTWAGTSFRR342 pKa = 4.48
Molecular weight: 36.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4197 |
342 |
3354 |
1399.0 |
156.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.1 ± 0.827 | 2.001 ± 0.634 |
6.338 ± 1.074 | 5.456 ± 0.777 |
4.265 ± 0.469 | 5.695 ± 1.243 |
2.43 ± 0.335 | 5.814 ± 0.312 |
5.575 ± 1.239 | 8.863 ± 1.442 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.359 ± 0.249 | 4.694 ± 0.318 |
5.337 ± 0.142 | 3.812 ± 0.858 |
5.218 ± 1.469 | 8.649 ± 2.546 |
6.004 ± 0.842 | 6.838 ± 0.636 |
1.287 ± 0.197 | 3.264 ± 0.856 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
