
Hubei tombus-like virus 42
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KG95|A0A1L3KG95_9VIRU Chropara_Vmeth domain-containing protein OS=Hubei tombus-like virus 42 OX=1923290 PE=4 SV=1
MM1 pKa = 7.83DD2 pKa = 5.74ASLFPLNIWIHH13 pKa = 6.3SSQQHH18 pKa = 4.18MNIRR22 pKa = 11.84RR23 pKa = 11.84AISHH27 pKa = 5.93TLVCVVQGCAVVKK40 pKa = 10.56DD41 pKa = 3.73RR42 pKa = 11.84LIAHH46 pKa = 6.23ITVNIDD52 pKa = 3.02DD53 pKa = 4.67PMPVVLQISPPSKK66 pKa = 9.03LQMAINHH73 pKa = 6.19ACNLRR78 pKa = 11.84SIVLSSTICVAKK90 pKa = 10.08RR91 pKa = 11.84LSGAISQAAQSIHH104 pKa = 6.25TGLHH108 pKa = 5.71LALHH112 pKa = 5.21YY113 pKa = 9.18TLLGAAVTAIIIWFIKK129 pKa = 8.16TRR131 pKa = 11.84KK132 pKa = 9.07CRR134 pKa = 11.84IIKK137 pKa = 9.22VRR139 pKa = 11.84NHH141 pKa = 5.44NQMADD146 pKa = 2.63IRR148 pKa = 11.84RR149 pKa = 11.84IKK151 pKa = 10.14EE152 pKa = 3.61QFATKK157 pKa = 10.01IKK159 pKa = 10.58RR160 pKa = 11.84LTPPEE165 pKa = 4.08FEE167 pKa = 4.11QGKK170 pKa = 8.91HH171 pKa = 5.81VICAWEE177 pKa = 4.08RR178 pKa = 11.84KK179 pKa = 5.45TTEE182 pKa = 3.74QIINEE187 pKa = 4.18WFHH190 pKa = 6.51DD191 pKa = 3.59QGIKK195 pKa = 10.46FRR197 pKa = 11.84DD198 pKa = 3.24IGGSRR203 pKa = 11.84VRR205 pKa = 11.84GSNYY209 pKa = 8.04ATSRR213 pKa = 11.84HH214 pKa = 5.4ICNPTLDD221 pKa = 3.61SSDD224 pKa = 2.75ILRR227 pKa = 11.84RR228 pKa = 11.84AKK230 pKa = 10.31QPTTIFSEE238 pKa = 5.01CTCLGQHH245 pKa = 6.32CPYY248 pKa = 10.29KK249 pKa = 10.66NEE251 pKa = 3.95YY252 pKa = 8.23PGAMFIHH259 pKa = 6.85SDD261 pKa = 3.89YY262 pKa = 11.47YY263 pKa = 11.5LDD265 pKa = 5.0ANALTDD271 pKa = 3.83CVRR274 pKa = 11.84SHH276 pKa = 6.11TFVVTHH282 pKa = 7.13RR283 pKa = 11.84FTGDD287 pKa = 2.45EE288 pKa = 3.98GTFFNEE294 pKa = 4.22AQWVKK299 pKa = 10.46HH300 pKa = 4.41GTKK303 pKa = 8.59ITMTTPDD310 pKa = 3.34GTKK313 pKa = 10.47YY314 pKa = 9.94NHH316 pKa = 7.59PYY318 pKa = 10.19NLWEE322 pKa = 4.15NEE324 pKa = 3.75GCIVGSSNACIYY336 pKa = 10.93AKK338 pKa = 10.5VGSTEE343 pKa = 3.86TSDD346 pKa = 3.2IYY348 pKa = 11.63YY349 pKa = 10.01LFPADD354 pKa = 3.56GTYY357 pKa = 10.95LKK359 pKa = 10.51DD360 pKa = 4.19DD361 pKa = 3.71RR362 pKa = 11.84LALKK366 pKa = 10.44RR367 pKa = 11.84STEE370 pKa = 3.99LAEE373 pKa = 4.27PLHH376 pKa = 6.07PSGYY380 pKa = 10.7VIIKK384 pKa = 9.63SDD386 pKa = 3.27QFFIYY391 pKa = 9.96DD392 pKa = 3.51DD393 pKa = 4.66HH394 pKa = 7.57DD395 pKa = 3.75NLMIKK400 pKa = 10.51GPVDD404 pKa = 3.67IISRR408 pKa = 11.84MKK410 pKa = 10.73LRR412 pKa = 11.84LGLAARR418 pKa = 11.84TSNFNSVFFSYY429 pKa = 10.55LQARR433 pKa = 11.84CQAEE437 pKa = 4.53EE438 pKa = 4.08IGITDD443 pKa = 4.4PLRR446 pKa = 11.84LTEE449 pKa = 3.9FALRR453 pKa = 11.84EE454 pKa = 4.17TEE456 pKa = 4.61QFALTIAPRR465 pKa = 11.84WSRR468 pKa = 11.84QWDD471 pKa = 3.73PANMTLCTGLKK482 pKa = 10.05LRR484 pKa = 11.84LHH486 pKa = 6.94RR487 pKa = 11.84MYY489 pKa = 10.39NTIIRR494 pKa = 11.84ATFDD498 pKa = 3.74LLPTCRR504 pKa = 11.84LRR506 pKa = 11.84CIFAHH511 pKa = 7.29RR512 pKa = 11.84ILAPWAYY519 pKa = 10.47RR520 pKa = 11.84EE521 pKa = 4.1ITLPGYY527 pKa = 10.31IMEE530 pKa = 4.65VRR532 pKa = 11.84PQHH535 pKa = 5.47TDD537 pKa = 2.73VRR539 pKa = 11.84GLRR542 pKa = 11.84PSGQPFPDD550 pKa = 4.97DD551 pKa = 3.6GTDD554 pKa = 3.68NDD556 pKa = 4.52AEE558 pKa = 4.48PDD560 pKa = 3.3NSEE563 pKa = 4.18SVRR566 pKa = 11.84ADD568 pKa = 3.41SVSGEE573 pKa = 4.22FDD575 pKa = 3.35SQSGNEE581 pKa = 3.83SDD583 pKa = 4.83GYY585 pKa = 8.99QTATSSRR592 pKa = 11.84SHH594 pKa = 5.87VRR596 pKa = 11.84NEE598 pKa = 3.67APIVDD603 pKa = 4.17TDD605 pKa = 3.65ASTTSTPGQSGRR617 pKa = 11.84TASLLPTDD625 pKa = 4.71GMDD628 pKa = 4.01SEE630 pKa = 4.97TSRR633 pKa = 11.84QIRR636 pKa = 11.84PITRR640 pKa = 11.84QDD642 pKa = 3.15SADD645 pKa = 3.65FEE647 pKa = 4.44KK648 pKa = 11.16SLIIFSGPSSSTSLPPTVVLDD669 pKa = 3.91PNRR672 pKa = 11.84NTVSSALYY680 pKa = 9.92DD681 pKa = 3.97DD682 pKa = 6.06PILLTAEE689 pKa = 4.38QFRR692 pKa = 11.84SLCQSKK698 pKa = 10.71DD699 pKa = 3.45AVLKK703 pKa = 10.84AHH705 pKa = 7.12FSTKK709 pKa = 7.85RR710 pKa = 11.84TRR712 pKa = 11.84EE713 pKa = 3.93QIYY716 pKa = 9.25HH717 pKa = 5.56QLVQEE722 pKa = 3.86ARR724 pKa = 11.84RR725 pKa = 11.84AKK727 pKa = 9.83RR728 pKa = 11.84HH729 pKa = 5.32PSEE732 pKa = 4.06TAGGGISEE740 pKa = 4.54DD741 pKa = 3.6PGNGVVPTPSRR752 pKa = 11.84TTTVRR757 pKa = 11.84PVGQEE762 pKa = 3.65IPKK765 pKa = 8.92MAPQTTRR772 pKa = 11.84DD773 pKa = 3.64SQNEE777 pKa = 4.24SGQSRR782 pKa = 11.84DD783 pKa = 3.27NVQRR787 pKa = 11.84RR788 pKa = 11.84PSSEE792 pKa = 3.6LHH794 pKa = 6.36KK795 pKa = 11.15ARR797 pKa = 11.84DD798 pKa = 3.8HH799 pKa = 6.15PQVHH803 pKa = 6.27RR804 pKa = 11.84PKK806 pKa = 10.27KK807 pKa = 8.4HH808 pKa = 5.49QSTHH812 pKa = 5.24RR813 pKa = 11.84RR814 pKa = 11.84VSGHH818 pKa = 5.66SRR820 pKa = 11.84SLHH823 pKa = 4.19QRR825 pKa = 11.84HH826 pKa = 5.94RR827 pKa = 11.84EE828 pKa = 3.94TRR830 pKa = 11.84SSRR833 pKa = 11.84PIPGQRR839 pKa = 11.84HH840 pKa = 4.36GAKK843 pKa = 10.15DD844 pKa = 3.3ARR846 pKa = 11.84KK847 pKa = 9.62EE848 pKa = 4.02DD849 pKa = 3.71GG850 pKa = 3.66
MM1 pKa = 7.83DD2 pKa = 5.74ASLFPLNIWIHH13 pKa = 6.3SSQQHH18 pKa = 4.18MNIRR22 pKa = 11.84RR23 pKa = 11.84AISHH27 pKa = 5.93TLVCVVQGCAVVKK40 pKa = 10.56DD41 pKa = 3.73RR42 pKa = 11.84LIAHH46 pKa = 6.23ITVNIDD52 pKa = 3.02DD53 pKa = 4.67PMPVVLQISPPSKK66 pKa = 9.03LQMAINHH73 pKa = 6.19ACNLRR78 pKa = 11.84SIVLSSTICVAKK90 pKa = 10.08RR91 pKa = 11.84LSGAISQAAQSIHH104 pKa = 6.25TGLHH108 pKa = 5.71LALHH112 pKa = 5.21YY113 pKa = 9.18TLLGAAVTAIIIWFIKK129 pKa = 8.16TRR131 pKa = 11.84KK132 pKa = 9.07CRR134 pKa = 11.84IIKK137 pKa = 9.22VRR139 pKa = 11.84NHH141 pKa = 5.44NQMADD146 pKa = 2.63IRR148 pKa = 11.84RR149 pKa = 11.84IKK151 pKa = 10.14EE152 pKa = 3.61QFATKK157 pKa = 10.01IKK159 pKa = 10.58RR160 pKa = 11.84LTPPEE165 pKa = 4.08FEE167 pKa = 4.11QGKK170 pKa = 8.91HH171 pKa = 5.81VICAWEE177 pKa = 4.08RR178 pKa = 11.84KK179 pKa = 5.45TTEE182 pKa = 3.74QIINEE187 pKa = 4.18WFHH190 pKa = 6.51DD191 pKa = 3.59QGIKK195 pKa = 10.46FRR197 pKa = 11.84DD198 pKa = 3.24IGGSRR203 pKa = 11.84VRR205 pKa = 11.84GSNYY209 pKa = 8.04ATSRR213 pKa = 11.84HH214 pKa = 5.4ICNPTLDD221 pKa = 3.61SSDD224 pKa = 2.75ILRR227 pKa = 11.84RR228 pKa = 11.84AKK230 pKa = 10.31QPTTIFSEE238 pKa = 5.01CTCLGQHH245 pKa = 6.32CPYY248 pKa = 10.29KK249 pKa = 10.66NEE251 pKa = 3.95YY252 pKa = 8.23PGAMFIHH259 pKa = 6.85SDD261 pKa = 3.89YY262 pKa = 11.47YY263 pKa = 11.5LDD265 pKa = 5.0ANALTDD271 pKa = 3.83CVRR274 pKa = 11.84SHH276 pKa = 6.11TFVVTHH282 pKa = 7.13RR283 pKa = 11.84FTGDD287 pKa = 2.45EE288 pKa = 3.98GTFFNEE294 pKa = 4.22AQWVKK299 pKa = 10.46HH300 pKa = 4.41GTKK303 pKa = 8.59ITMTTPDD310 pKa = 3.34GTKK313 pKa = 10.47YY314 pKa = 9.94NHH316 pKa = 7.59PYY318 pKa = 10.19NLWEE322 pKa = 4.15NEE324 pKa = 3.75GCIVGSSNACIYY336 pKa = 10.93AKK338 pKa = 10.5VGSTEE343 pKa = 3.86TSDD346 pKa = 3.2IYY348 pKa = 11.63YY349 pKa = 10.01LFPADD354 pKa = 3.56GTYY357 pKa = 10.95LKK359 pKa = 10.51DD360 pKa = 4.19DD361 pKa = 3.71RR362 pKa = 11.84LALKK366 pKa = 10.44RR367 pKa = 11.84STEE370 pKa = 3.99LAEE373 pKa = 4.27PLHH376 pKa = 6.07PSGYY380 pKa = 10.7VIIKK384 pKa = 9.63SDD386 pKa = 3.27QFFIYY391 pKa = 9.96DD392 pKa = 3.51DD393 pKa = 4.66HH394 pKa = 7.57DD395 pKa = 3.75NLMIKK400 pKa = 10.51GPVDD404 pKa = 3.67IISRR408 pKa = 11.84MKK410 pKa = 10.73LRR412 pKa = 11.84LGLAARR418 pKa = 11.84TSNFNSVFFSYY429 pKa = 10.55LQARR433 pKa = 11.84CQAEE437 pKa = 4.53EE438 pKa = 4.08IGITDD443 pKa = 4.4PLRR446 pKa = 11.84LTEE449 pKa = 3.9FALRR453 pKa = 11.84EE454 pKa = 4.17TEE456 pKa = 4.61QFALTIAPRR465 pKa = 11.84WSRR468 pKa = 11.84QWDD471 pKa = 3.73PANMTLCTGLKK482 pKa = 10.05LRR484 pKa = 11.84LHH486 pKa = 6.94RR487 pKa = 11.84MYY489 pKa = 10.39NTIIRR494 pKa = 11.84ATFDD498 pKa = 3.74LLPTCRR504 pKa = 11.84LRR506 pKa = 11.84CIFAHH511 pKa = 7.29RR512 pKa = 11.84ILAPWAYY519 pKa = 10.47RR520 pKa = 11.84EE521 pKa = 4.1ITLPGYY527 pKa = 10.31IMEE530 pKa = 4.65VRR532 pKa = 11.84PQHH535 pKa = 5.47TDD537 pKa = 2.73VRR539 pKa = 11.84GLRR542 pKa = 11.84PSGQPFPDD550 pKa = 4.97DD551 pKa = 3.6GTDD554 pKa = 3.68NDD556 pKa = 4.52AEE558 pKa = 4.48PDD560 pKa = 3.3NSEE563 pKa = 4.18SVRR566 pKa = 11.84ADD568 pKa = 3.41SVSGEE573 pKa = 4.22FDD575 pKa = 3.35SQSGNEE581 pKa = 3.83SDD583 pKa = 4.83GYY585 pKa = 8.99QTATSSRR592 pKa = 11.84SHH594 pKa = 5.87VRR596 pKa = 11.84NEE598 pKa = 3.67APIVDD603 pKa = 4.17TDD605 pKa = 3.65ASTTSTPGQSGRR617 pKa = 11.84TASLLPTDD625 pKa = 4.71GMDD628 pKa = 4.01SEE630 pKa = 4.97TSRR633 pKa = 11.84QIRR636 pKa = 11.84PITRR640 pKa = 11.84QDD642 pKa = 3.15SADD645 pKa = 3.65FEE647 pKa = 4.44KK648 pKa = 11.16SLIIFSGPSSSTSLPPTVVLDD669 pKa = 3.91PNRR672 pKa = 11.84NTVSSALYY680 pKa = 9.92DD681 pKa = 3.97DD682 pKa = 6.06PILLTAEE689 pKa = 4.38QFRR692 pKa = 11.84SLCQSKK698 pKa = 10.71DD699 pKa = 3.45AVLKK703 pKa = 10.84AHH705 pKa = 7.12FSTKK709 pKa = 7.85RR710 pKa = 11.84TRR712 pKa = 11.84EE713 pKa = 3.93QIYY716 pKa = 9.25HH717 pKa = 5.56QLVQEE722 pKa = 3.86ARR724 pKa = 11.84RR725 pKa = 11.84AKK727 pKa = 9.83RR728 pKa = 11.84HH729 pKa = 5.32PSEE732 pKa = 4.06TAGGGISEE740 pKa = 4.54DD741 pKa = 3.6PGNGVVPTPSRR752 pKa = 11.84TTTVRR757 pKa = 11.84PVGQEE762 pKa = 3.65IPKK765 pKa = 8.92MAPQTTRR772 pKa = 11.84DD773 pKa = 3.64SQNEE777 pKa = 4.24SGQSRR782 pKa = 11.84DD783 pKa = 3.27NVQRR787 pKa = 11.84RR788 pKa = 11.84PSSEE792 pKa = 3.6LHH794 pKa = 6.36KK795 pKa = 11.15ARR797 pKa = 11.84DD798 pKa = 3.8HH799 pKa = 6.15PQVHH803 pKa = 6.27RR804 pKa = 11.84PKK806 pKa = 10.27KK807 pKa = 8.4HH808 pKa = 5.49QSTHH812 pKa = 5.24RR813 pKa = 11.84RR814 pKa = 11.84VSGHH818 pKa = 5.66SRR820 pKa = 11.84SLHH823 pKa = 4.19QRR825 pKa = 11.84HH826 pKa = 5.94RR827 pKa = 11.84EE828 pKa = 3.94TRR830 pKa = 11.84SSRR833 pKa = 11.84PIPGQRR839 pKa = 11.84HH840 pKa = 4.36GAKK843 pKa = 10.15DD844 pKa = 3.3ARR846 pKa = 11.84KK847 pKa = 9.62EE848 pKa = 4.02DD849 pKa = 3.71GG850 pKa = 3.66
Molecular weight: 95.58 kDa
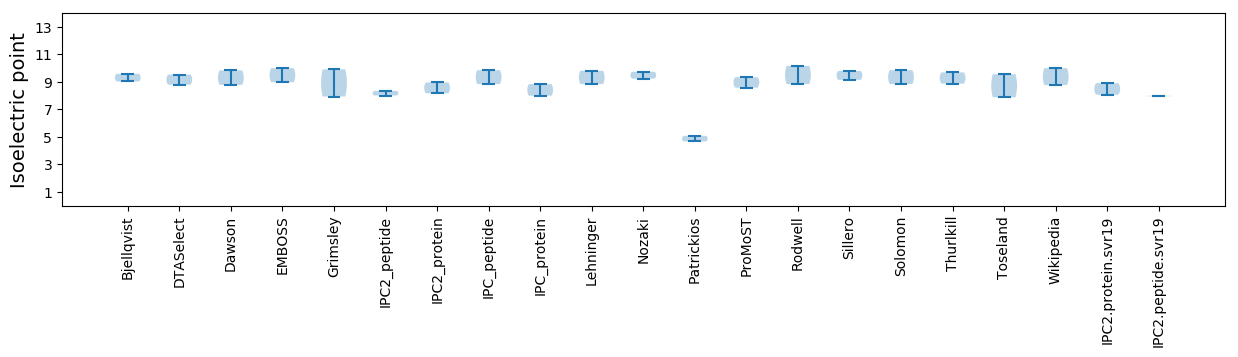
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG95|A0A1L3KG95_9VIRU Chropara_Vmeth domain-containing protein OS=Hubei tombus-like virus 42 OX=1923290 PE=4 SV=1
MM1 pKa = 7.21FVDD4 pKa = 4.7CGRR7 pKa = 11.84VDD9 pKa = 3.5SPFRR13 pKa = 11.84TTARR17 pKa = 11.84IMMPNLTTANLSVPIVCQEE36 pKa = 3.7NLIASLEE43 pKa = 4.18TRR45 pKa = 11.84VMGTKK50 pKa = 9.37PQPHH54 pKa = 7.35PGLTFAMKK62 pKa = 9.49PQLLIPMPLLPQHH75 pKa = 6.9PANQDD80 pKa = 2.87GLLAYY85 pKa = 7.94CQRR88 pKa = 11.84MGWTAKK94 pKa = 10.6LVDD97 pKa = 4.87KK98 pKa = 10.98YY99 pKa = 11.43DD100 pKa = 3.87PSQDD104 pKa = 3.54KK105 pKa = 10.41IVQILRR111 pKa = 11.84NPSSYY116 pKa = 11.1SADD119 pKa = 3.38PALAPLCHH127 pKa = 6.08QRR129 pKa = 11.84SYY131 pKa = 8.08WTPTEE136 pKa = 3.91IPLALHH142 pKa = 5.62YY143 pKa = 7.36TTTLYY148 pKa = 10.83SSQPSSLDD156 pKa = 2.94HH157 pKa = 6.13CVNRR161 pKa = 11.84KK162 pKa = 6.86TLSSKK167 pKa = 9.43LTSPPNGLEE176 pKa = 3.76NKK178 pKa = 8.64FTINSSKK185 pKa = 10.74KK186 pKa = 8.98HH187 pKa = 4.85VEE189 pKa = 3.89PRR191 pKa = 11.84DD192 pKa = 3.51TPLRR196 pKa = 11.84LLVEE200 pKa = 4.62EE201 pKa = 4.38FLKK204 pKa = 10.03TPEE207 pKa = 4.32MEE209 pKa = 4.29WFPHH213 pKa = 5.97HH214 pKa = 7.15LEE216 pKa = 4.63PLPFDD221 pKa = 3.46QWVKK225 pKa = 10.68RR226 pKa = 11.84YY227 pKa = 8.37PKK229 pKa = 9.34WRR231 pKa = 11.84RR232 pKa = 11.84KK233 pKa = 7.55QLEE236 pKa = 3.94IAKK239 pKa = 9.32MKK241 pKa = 9.67VDD243 pKa = 3.45KK244 pKa = 11.42VGITSKK250 pKa = 10.67DD251 pKa = 2.99ARR253 pKa = 11.84VRR255 pKa = 11.84NFIKK259 pKa = 10.65RR260 pKa = 11.84EE261 pKa = 4.11TTHH264 pKa = 7.13KK265 pKa = 10.12FTDD268 pKa = 3.55PRR270 pKa = 11.84NISPRR275 pKa = 11.84TDD277 pKa = 2.96EE278 pKa = 4.29FLVTVGPYY286 pKa = 9.37ISAIEE291 pKa = 4.09KK292 pKa = 9.31HH293 pKa = 5.64AHH295 pKa = 5.69HH296 pKa = 7.04APFLVKK302 pKa = 10.55GMGLKK307 pKa = 9.04MRR309 pKa = 11.84AKK311 pKa = 10.72KK312 pKa = 9.34MDD314 pKa = 3.46KK315 pKa = 10.43LLGFGRR321 pKa = 11.84YY322 pKa = 9.43LEE324 pKa = 4.2VDD326 pKa = 4.43FTRR329 pKa = 11.84FDD331 pKa = 3.46KK332 pKa = 11.04TISADD337 pKa = 3.1IIRR340 pKa = 11.84IVEE343 pKa = 4.27KK344 pKa = 11.07YY345 pKa = 10.69LLTKK349 pKa = 10.05PYY351 pKa = 10.74SRR353 pKa = 11.84DD354 pKa = 2.52HH355 pKa = 6.41HH356 pKa = 7.1AYY358 pKa = 8.18HH359 pKa = 6.29QCMKK363 pKa = 10.65QLTTTSGVSYY373 pKa = 10.45FGTRR377 pKa = 11.84YY378 pKa = 9.08KK379 pKa = 11.1VKK381 pKa = 9.35GTRR384 pKa = 11.84CSGDD388 pKa = 2.7AHH390 pKa = 6.14TSLGNGLINRR400 pKa = 11.84FIIWLCLRR408 pKa = 11.84KK409 pKa = 9.8LPQNAWDD416 pKa = 4.09SMHH419 pKa = 6.9EE420 pKa = 4.16GDD422 pKa = 5.56DD423 pKa = 4.41GIIGVSLSYY432 pKa = 11.23VNQAVYY438 pKa = 10.86NLQFLACLGFEE449 pKa = 4.64SKK451 pKa = 10.96LKK453 pKa = 8.54VCKK456 pKa = 10.46SIEE459 pKa = 3.96EE460 pKa = 4.2VVFCGRR466 pKa = 11.84RR467 pKa = 11.84HH468 pKa = 4.7VRR470 pKa = 11.84TAKK473 pKa = 10.34EE474 pKa = 3.76SATICDD480 pKa = 4.12VIRR483 pKa = 11.84TMRR486 pKa = 11.84KK487 pKa = 9.22FNTTCSLGPPDD498 pKa = 4.42LLLYY502 pKa = 10.88AKK504 pKa = 10.12ALSYY508 pKa = 11.18NYY510 pKa = 9.6TDD512 pKa = 4.21ADD514 pKa = 3.95TPIIGPVSYY523 pKa = 10.96AVARR527 pKa = 11.84CLAHH531 pKa = 7.1CNIKK535 pKa = 10.52YY536 pKa = 9.98SRR538 pKa = 11.84KK539 pKa = 7.68QFKK542 pKa = 10.09RR543 pKa = 11.84ALKK546 pKa = 8.84TAVLEE551 pKa = 4.12RR552 pKa = 11.84WVLNDD557 pKa = 3.48NIGSISYY564 pKa = 9.96HH565 pKa = 6.22RR566 pKa = 11.84LLNAKK571 pKa = 9.46PPDD574 pKa = 3.4ISPYY578 pKa = 10.54LITASLMLNRR588 pKa = 11.84PTSVQKK594 pKa = 10.97LMQQQ598 pKa = 3.14
MM1 pKa = 7.21FVDD4 pKa = 4.7CGRR7 pKa = 11.84VDD9 pKa = 3.5SPFRR13 pKa = 11.84TTARR17 pKa = 11.84IMMPNLTTANLSVPIVCQEE36 pKa = 3.7NLIASLEE43 pKa = 4.18TRR45 pKa = 11.84VMGTKK50 pKa = 9.37PQPHH54 pKa = 7.35PGLTFAMKK62 pKa = 9.49PQLLIPMPLLPQHH75 pKa = 6.9PANQDD80 pKa = 2.87GLLAYY85 pKa = 7.94CQRR88 pKa = 11.84MGWTAKK94 pKa = 10.6LVDD97 pKa = 4.87KK98 pKa = 10.98YY99 pKa = 11.43DD100 pKa = 3.87PSQDD104 pKa = 3.54KK105 pKa = 10.41IVQILRR111 pKa = 11.84NPSSYY116 pKa = 11.1SADD119 pKa = 3.38PALAPLCHH127 pKa = 6.08QRR129 pKa = 11.84SYY131 pKa = 8.08WTPTEE136 pKa = 3.91IPLALHH142 pKa = 5.62YY143 pKa = 7.36TTTLYY148 pKa = 10.83SSQPSSLDD156 pKa = 2.94HH157 pKa = 6.13CVNRR161 pKa = 11.84KK162 pKa = 6.86TLSSKK167 pKa = 9.43LTSPPNGLEE176 pKa = 3.76NKK178 pKa = 8.64FTINSSKK185 pKa = 10.74KK186 pKa = 8.98HH187 pKa = 4.85VEE189 pKa = 3.89PRR191 pKa = 11.84DD192 pKa = 3.51TPLRR196 pKa = 11.84LLVEE200 pKa = 4.62EE201 pKa = 4.38FLKK204 pKa = 10.03TPEE207 pKa = 4.32MEE209 pKa = 4.29WFPHH213 pKa = 5.97HH214 pKa = 7.15LEE216 pKa = 4.63PLPFDD221 pKa = 3.46QWVKK225 pKa = 10.68RR226 pKa = 11.84YY227 pKa = 8.37PKK229 pKa = 9.34WRR231 pKa = 11.84RR232 pKa = 11.84KK233 pKa = 7.55QLEE236 pKa = 3.94IAKK239 pKa = 9.32MKK241 pKa = 9.67VDD243 pKa = 3.45KK244 pKa = 11.42VGITSKK250 pKa = 10.67DD251 pKa = 2.99ARR253 pKa = 11.84VRR255 pKa = 11.84NFIKK259 pKa = 10.65RR260 pKa = 11.84EE261 pKa = 4.11TTHH264 pKa = 7.13KK265 pKa = 10.12FTDD268 pKa = 3.55PRR270 pKa = 11.84NISPRR275 pKa = 11.84TDD277 pKa = 2.96EE278 pKa = 4.29FLVTVGPYY286 pKa = 9.37ISAIEE291 pKa = 4.09KK292 pKa = 9.31HH293 pKa = 5.64AHH295 pKa = 5.69HH296 pKa = 7.04APFLVKK302 pKa = 10.55GMGLKK307 pKa = 9.04MRR309 pKa = 11.84AKK311 pKa = 10.72KK312 pKa = 9.34MDD314 pKa = 3.46KK315 pKa = 10.43LLGFGRR321 pKa = 11.84YY322 pKa = 9.43LEE324 pKa = 4.2VDD326 pKa = 4.43FTRR329 pKa = 11.84FDD331 pKa = 3.46KK332 pKa = 11.04TISADD337 pKa = 3.1IIRR340 pKa = 11.84IVEE343 pKa = 4.27KK344 pKa = 11.07YY345 pKa = 10.69LLTKK349 pKa = 10.05PYY351 pKa = 10.74SRR353 pKa = 11.84DD354 pKa = 2.52HH355 pKa = 6.41HH356 pKa = 7.1AYY358 pKa = 8.18HH359 pKa = 6.29QCMKK363 pKa = 10.65QLTTTSGVSYY373 pKa = 10.45FGTRR377 pKa = 11.84YY378 pKa = 9.08KK379 pKa = 11.1VKK381 pKa = 9.35GTRR384 pKa = 11.84CSGDD388 pKa = 2.7AHH390 pKa = 6.14TSLGNGLINRR400 pKa = 11.84FIIWLCLRR408 pKa = 11.84KK409 pKa = 9.8LPQNAWDD416 pKa = 4.09SMHH419 pKa = 6.9EE420 pKa = 4.16GDD422 pKa = 5.56DD423 pKa = 4.41GIIGVSLSYY432 pKa = 11.23VNQAVYY438 pKa = 10.86NLQFLACLGFEE449 pKa = 4.64SKK451 pKa = 10.96LKK453 pKa = 8.54VCKK456 pKa = 10.46SIEE459 pKa = 3.96EE460 pKa = 4.2VVFCGRR466 pKa = 11.84RR467 pKa = 11.84HH468 pKa = 4.7VRR470 pKa = 11.84TAKK473 pKa = 10.34EE474 pKa = 3.76SATICDD480 pKa = 4.12VIRR483 pKa = 11.84TMRR486 pKa = 11.84KK487 pKa = 9.22FNTTCSLGPPDD498 pKa = 4.42LLLYY502 pKa = 10.88AKK504 pKa = 10.12ALSYY508 pKa = 11.18NYY510 pKa = 9.6TDD512 pKa = 4.21ADD514 pKa = 3.95TPIIGPVSYY523 pKa = 10.96AVARR527 pKa = 11.84CLAHH531 pKa = 7.1CNIKK535 pKa = 10.52YY536 pKa = 9.98SRR538 pKa = 11.84KK539 pKa = 7.68QFKK542 pKa = 10.09RR543 pKa = 11.84ALKK546 pKa = 8.84TAVLEE551 pKa = 4.12RR552 pKa = 11.84WVLNDD557 pKa = 3.48NIGSISYY564 pKa = 9.96HH565 pKa = 6.22RR566 pKa = 11.84LLNAKK571 pKa = 9.46PPDD574 pKa = 3.4ISPYY578 pKa = 10.54LITASLMLNRR588 pKa = 11.84PTSVQKK594 pKa = 10.97LMQQQ598 pKa = 3.14
Molecular weight: 68.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1448 |
598 |
850 |
724.0 |
81.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.423 ± 0.345 | 2.279 ± 0.139 |
5.525 ± 0.409 | 4.213 ± 0.324 |
3.384 ± 0.077 | 4.972 ± 0.379 |
3.867 ± 0.317 | 6.423 ± 0.548 |
5.525 ± 1.213 | 8.425 ± 1.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.425 | 3.729 ± 0.071 |
6.215 ± 0.388 | 4.282 ± 0.467 |
7.528 ± 0.711 | 8.011 ± 0.7 |
7.666 ± 0.288 | 5.18 ± 0.307 |
1.174 ± 0.099 | 3.039 ± 0.489 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
