
Kanyawara virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Kanyawara ledantevirus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
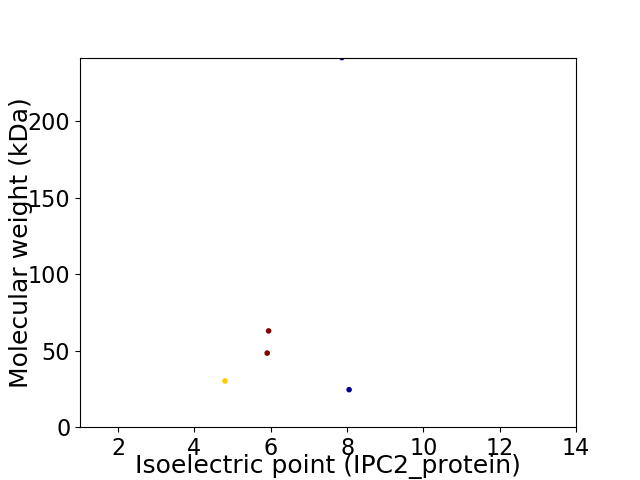
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
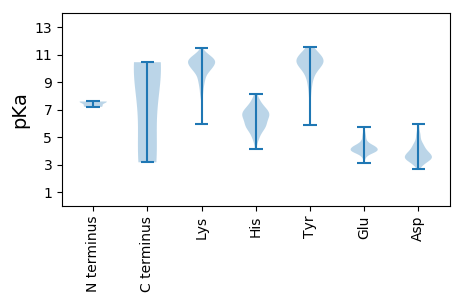
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A220FHW5|A0A220FHW5_9RHAB Phosphoprotein OS=Kanyawara virus OX=2014931 PE=4 SV=1
MM1 pKa = 7.59EE2 pKa = 5.27FNRR5 pKa = 11.84SKK7 pKa = 10.22IHH9 pKa = 5.85SAASRR14 pKa = 11.84LSWSAVNTNLGGLEE28 pKa = 3.89EE29 pKa = 4.54DD30 pKa = 3.51AEE32 pKa = 4.44EE33 pKa = 4.85IILEE37 pKa = 4.13NSSSVKK43 pKa = 10.36EE44 pKa = 4.2ADD46 pKa = 3.14QSAYY50 pKa = 10.9SSWANNPVIEE60 pKa = 4.3TLDD63 pKa = 3.83EE64 pKa = 4.37IFSEE68 pKa = 5.5DD69 pKa = 4.21SDD71 pKa = 3.94TDD73 pKa = 3.72TEE75 pKa = 4.44KK76 pKa = 10.9QSSSQNSKK84 pKa = 8.02STEE87 pKa = 4.15DD88 pKa = 3.5IVCKK92 pKa = 10.3EE93 pKa = 4.29SNSTEE98 pKa = 4.01DD99 pKa = 3.64VIEE102 pKa = 4.14SLKK105 pKa = 11.05KK106 pKa = 9.12EE107 pKa = 4.36DD108 pKa = 3.18EE109 pKa = 4.05RR110 pKa = 11.84QRR112 pKa = 11.84AGKK115 pKa = 9.39KK116 pKa = 9.51DD117 pKa = 3.33KK118 pKa = 11.02RR119 pKa = 11.84SITLIIPNISRR130 pKa = 11.84IHH132 pKa = 5.11NQRR135 pKa = 11.84MLDD138 pKa = 3.78DD139 pKa = 4.47VIKK142 pKa = 10.61EE143 pKa = 3.78ITNNLVEE150 pKa = 4.17QLGFQTYY157 pKa = 9.45PDD159 pKa = 3.56LMEE162 pKa = 5.71INDD165 pKa = 4.51RR166 pKa = 11.84SLKK169 pKa = 10.65LYY171 pKa = 10.67VSADD175 pKa = 3.22NDD177 pKa = 3.71VATSASKK184 pKa = 11.02VSLDD188 pKa = 3.61SKK190 pKa = 9.7PTISTLPQNSPEE202 pKa = 4.02SLIDD206 pKa = 3.45RR207 pKa = 11.84FRR209 pKa = 11.84TGITFQKK216 pKa = 10.37RR217 pKa = 11.84RR218 pKa = 11.84GGDD221 pKa = 3.35LKK223 pKa = 10.17ITLSSGRR230 pKa = 11.84ITEE233 pKa = 5.24LMILDD238 pKa = 5.17CYY240 pKa = 10.66SAAANEE246 pKa = 4.21DD247 pKa = 3.68DD248 pKa = 5.8AIRR251 pKa = 11.84TILKK255 pKa = 9.77KK256 pKa = 10.83ANLYY260 pKa = 7.05TTLTLLTNYY269 pKa = 9.94KK270 pKa = 10.44
MM1 pKa = 7.59EE2 pKa = 5.27FNRR5 pKa = 11.84SKK7 pKa = 10.22IHH9 pKa = 5.85SAASRR14 pKa = 11.84LSWSAVNTNLGGLEE28 pKa = 3.89EE29 pKa = 4.54DD30 pKa = 3.51AEE32 pKa = 4.44EE33 pKa = 4.85IILEE37 pKa = 4.13NSSSVKK43 pKa = 10.36EE44 pKa = 4.2ADD46 pKa = 3.14QSAYY50 pKa = 10.9SSWANNPVIEE60 pKa = 4.3TLDD63 pKa = 3.83EE64 pKa = 4.37IFSEE68 pKa = 5.5DD69 pKa = 4.21SDD71 pKa = 3.94TDD73 pKa = 3.72TEE75 pKa = 4.44KK76 pKa = 10.9QSSSQNSKK84 pKa = 8.02STEE87 pKa = 4.15DD88 pKa = 3.5IVCKK92 pKa = 10.3EE93 pKa = 4.29SNSTEE98 pKa = 4.01DD99 pKa = 3.64VIEE102 pKa = 4.14SLKK105 pKa = 11.05KK106 pKa = 9.12EE107 pKa = 4.36DD108 pKa = 3.18EE109 pKa = 4.05RR110 pKa = 11.84QRR112 pKa = 11.84AGKK115 pKa = 9.39KK116 pKa = 9.51DD117 pKa = 3.33KK118 pKa = 11.02RR119 pKa = 11.84SITLIIPNISRR130 pKa = 11.84IHH132 pKa = 5.11NQRR135 pKa = 11.84MLDD138 pKa = 3.78DD139 pKa = 4.47VIKK142 pKa = 10.61EE143 pKa = 3.78ITNNLVEE150 pKa = 4.17QLGFQTYY157 pKa = 9.45PDD159 pKa = 3.56LMEE162 pKa = 5.71INDD165 pKa = 4.51RR166 pKa = 11.84SLKK169 pKa = 10.65LYY171 pKa = 10.67VSADD175 pKa = 3.22NDD177 pKa = 3.71VATSASKK184 pKa = 11.02VSLDD188 pKa = 3.61SKK190 pKa = 9.7PTISTLPQNSPEE202 pKa = 4.02SLIDD206 pKa = 3.45RR207 pKa = 11.84FRR209 pKa = 11.84TGITFQKK216 pKa = 10.37RR217 pKa = 11.84RR218 pKa = 11.84GGDD221 pKa = 3.35LKK223 pKa = 10.17ITLSSGRR230 pKa = 11.84ITEE233 pKa = 5.24LMILDD238 pKa = 5.17CYY240 pKa = 10.66SAAANEE246 pKa = 4.21DD247 pKa = 3.68DD248 pKa = 5.8AIRR251 pKa = 11.84TILKK255 pKa = 9.77KK256 pKa = 10.83ANLYY260 pKa = 7.05TTLTLLTNYY269 pKa = 9.94KK270 pKa = 10.44
Molecular weight: 30.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220FHW5|A0A220FHW5_9RHAB Phosphoprotein OS=Kanyawara virus OX=2014931 PE=4 SV=1
MM1 pKa = 7.26LQRR4 pKa = 11.84FKK6 pKa = 11.24KK7 pKa = 8.16HH8 pKa = 5.69TKK10 pKa = 10.14LKK12 pKa = 9.2TPSAPSQSTIYY23 pKa = 10.81GLSTKK28 pKa = 9.46WDD30 pKa = 3.53LHH32 pKa = 5.67TNPQIDD38 pKa = 3.84NVVQPYY44 pKa = 9.46RR45 pKa = 11.84VEE47 pKa = 3.8EE48 pKa = 3.92TLRR51 pKa = 11.84IQNKK55 pKa = 9.79LEE57 pKa = 3.82IRR59 pKa = 11.84AKK61 pKa = 10.53EE62 pKa = 4.02PLRR65 pKa = 11.84DD66 pKa = 3.54LTDD69 pKa = 3.63CLKK72 pKa = 10.51IAEE75 pKa = 4.23VWIDD79 pKa = 3.85EE80 pKa = 4.32QNCPYY85 pKa = 8.96WQVGIDD91 pKa = 3.3TWIFICMALHH101 pKa = 5.53AQKK104 pKa = 10.69DD105 pKa = 4.29GSCRR109 pKa = 11.84YY110 pKa = 7.88IHH112 pKa = 7.65LYY114 pKa = 8.42RR115 pKa = 11.84TQLDD119 pKa = 3.45QVVDD123 pKa = 4.85FKK125 pKa = 11.0IQRR128 pKa = 11.84PFLEE132 pKa = 4.67RR133 pKa = 11.84INYY136 pKa = 7.89KK137 pKa = 10.02KK138 pKa = 9.98HH139 pKa = 4.3SQYY142 pKa = 11.45YY143 pKa = 6.31EE144 pKa = 3.96TTHH147 pKa = 6.82RR148 pKa = 11.84GSTCSVSYY156 pKa = 10.37EE157 pKa = 4.32SNLALSKK164 pKa = 10.1RR165 pKa = 11.84TGIPSHH171 pKa = 6.47VLYY174 pKa = 10.41QYY176 pKa = 10.11PLKK179 pKa = 10.93DD180 pKa = 3.47GNSPPKK186 pKa = 10.37FDD188 pKa = 4.93DD189 pKa = 4.59LGIKK193 pKa = 8.81TPIVVTYY200 pKa = 10.66ADD202 pKa = 5.06DD203 pKa = 4.32GSQIIGTKK211 pKa = 9.93
MM1 pKa = 7.26LQRR4 pKa = 11.84FKK6 pKa = 11.24KK7 pKa = 8.16HH8 pKa = 5.69TKK10 pKa = 10.14LKK12 pKa = 9.2TPSAPSQSTIYY23 pKa = 10.81GLSTKK28 pKa = 9.46WDD30 pKa = 3.53LHH32 pKa = 5.67TNPQIDD38 pKa = 3.84NVVQPYY44 pKa = 9.46RR45 pKa = 11.84VEE47 pKa = 3.8EE48 pKa = 3.92TLRR51 pKa = 11.84IQNKK55 pKa = 9.79LEE57 pKa = 3.82IRR59 pKa = 11.84AKK61 pKa = 10.53EE62 pKa = 4.02PLRR65 pKa = 11.84DD66 pKa = 3.54LTDD69 pKa = 3.63CLKK72 pKa = 10.51IAEE75 pKa = 4.23VWIDD79 pKa = 3.85EE80 pKa = 4.32QNCPYY85 pKa = 8.96WQVGIDD91 pKa = 3.3TWIFICMALHH101 pKa = 5.53AQKK104 pKa = 10.69DD105 pKa = 4.29GSCRR109 pKa = 11.84YY110 pKa = 7.88IHH112 pKa = 7.65LYY114 pKa = 8.42RR115 pKa = 11.84TQLDD119 pKa = 3.45QVVDD123 pKa = 4.85FKK125 pKa = 11.0IQRR128 pKa = 11.84PFLEE132 pKa = 4.67RR133 pKa = 11.84INYY136 pKa = 7.89KK137 pKa = 10.02KK138 pKa = 9.98HH139 pKa = 4.3SQYY142 pKa = 11.45YY143 pKa = 6.31EE144 pKa = 3.96TTHH147 pKa = 6.82RR148 pKa = 11.84GSTCSVSYY156 pKa = 10.37EE157 pKa = 4.32SNLALSKK164 pKa = 10.1RR165 pKa = 11.84TGIPSHH171 pKa = 6.47VLYY174 pKa = 10.41QYY176 pKa = 10.11PLKK179 pKa = 10.93DD180 pKa = 3.47GNSPPKK186 pKa = 10.37FDD188 pKa = 4.93DD189 pKa = 4.59LGIKK193 pKa = 8.81TPIVVTYY200 pKa = 10.66ADD202 pKa = 5.06DD203 pKa = 4.32GSQIIGTKK211 pKa = 9.93
Molecular weight: 24.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3573 |
211 |
2117 |
714.6 |
81.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.618 ± 0.435 | 1.959 ± 0.361 |
5.682 ± 0.5 | 5.57 ± 0.676 |
3.834 ± 0.463 | 5.43 ± 0.589 |
2.883 ± 0.449 | 7.529 ± 0.582 |
6.465 ± 0.282 | 10.327 ± 0.652 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.295 ± 0.276 | 5.43 ± 0.348 |
4.086 ± 0.365 | 3.61 ± 0.481 |
5.038 ± 0.337 | 8.34 ± 0.89 |
6.297 ± 0.572 | 5.038 ± 0.419 |
1.931 ± 0.228 | 3.638 ± 0.352 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
