
Desulfovibrio sp. X2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio; unclassified Desulfovibrio
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
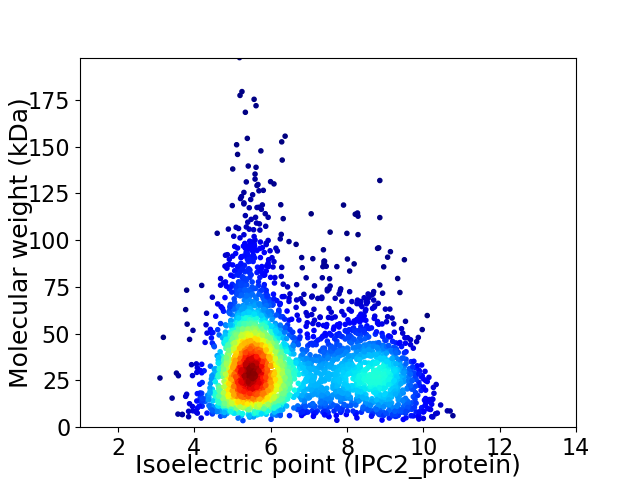
Virtual 2D-PAGE plot for 3444 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S7VAY5|S7VAY5_9DELT Uncharacterized protein OS=Desulfovibrio sp. X2 OX=941449 GN=dsx2_0088 PE=4 SV=1
MM1 pKa = 7.5SLTGSLYY8 pKa = 10.86AGISGLQAHH17 pKa = 5.6SQKK20 pKa = 10.22MSVIGNNLANTSTIGFKK37 pKa = 10.64SSTMQFEE44 pKa = 5.14DD45 pKa = 3.43IFYY48 pKa = 10.69QSVNTGAGIGQVGVGAGVAAIYY70 pKa = 11.02SNFAQGSYY78 pKa = 10.48EE79 pKa = 4.24STSEE83 pKa = 4.03ATDD86 pKa = 3.38VAIGGTGFFMVNNPNTDD103 pKa = 2.65EE104 pKa = 4.1MYY106 pKa = 8.9YY107 pKa = 9.98TRR109 pKa = 11.84AGEE112 pKa = 3.95FRR114 pKa = 11.84FDD116 pKa = 3.23NSGYY120 pKa = 10.78LVDD123 pKa = 3.78TNGYY127 pKa = 8.89RR128 pKa = 11.84VQGWQVKK135 pKa = 9.81DD136 pKa = 3.87GSSSGTVQTTGVSGDD151 pKa = 3.32IQLKK155 pKa = 9.89NFQSPPEE162 pKa = 4.13ATSTVSMLLNLDD174 pKa = 3.79SSSDD178 pKa = 3.74DD179 pKa = 3.94DD180 pKa = 4.28SANAANPFFSLFGNWDD196 pKa = 3.49GTDD199 pKa = 3.37SDD201 pKa = 4.44TPIADD206 pKa = 3.35SRR208 pKa = 11.84YY209 pKa = 9.74AYY211 pKa = 10.69QNTITVYY218 pKa = 10.78DD219 pKa = 4.2EE220 pKa = 4.38NGSAQTLTVYY230 pKa = 10.72CDD232 pKa = 3.53PVKK235 pKa = 10.71DD236 pKa = 3.7ASVVSNSGGNLQWEE250 pKa = 5.19FIVACDD256 pKa = 3.57PSEE259 pKa = 4.43DD260 pKa = 3.3GRR262 pKa = 11.84TIGGQKK268 pKa = 10.1VGSTSAAGLLMTGTLTFNAAGQLTGMSAYY297 pKa = 8.93TLASGASGDD306 pKa = 5.44LKK308 pKa = 11.14DD309 pKa = 4.11LSNWTTADD317 pKa = 4.0LNDD320 pKa = 4.93DD321 pKa = 3.92GLPTFTANFTGEE333 pKa = 4.03ADD335 pKa = 3.36ASATGEE341 pKa = 4.14ANASAIAIDD350 pKa = 4.69FGIHH354 pKa = 7.31DD355 pKa = 4.7SGSPTSWSGSASNASMVGNNPSMLDD380 pKa = 3.15NFTSPKK386 pKa = 9.91ISALTTTSYY395 pKa = 11.29DD396 pKa = 3.4SSSTTISQSQDD407 pKa = 2.54GYY409 pKa = 11.72GPGFLTDD416 pKa = 3.37ISVNRR421 pKa = 11.84DD422 pKa = 2.85GVITGTYY429 pKa = 10.73SNGQVLDD436 pKa = 4.39LYY438 pKa = 11.38VLTLASFTNPYY449 pKa = 9.8GLSRR453 pKa = 11.84EE454 pKa = 3.92GDD456 pKa = 3.55NLFSATRR463 pKa = 11.84EE464 pKa = 3.79SGAAVTGTANTGQLGSISSNTLEE487 pKa = 4.14QSNVDD492 pKa = 3.52TATEE496 pKa = 4.09MVDD499 pKa = 5.24LITTQRR505 pKa = 11.84GFEE508 pKa = 4.22ANSKK512 pKa = 10.68VITTADD518 pKa = 3.25SMLSEE523 pKa = 5.48LIQLKK528 pKa = 10.17RR529 pKa = 3.41
MM1 pKa = 7.5SLTGSLYY8 pKa = 10.86AGISGLQAHH17 pKa = 5.6SQKK20 pKa = 10.22MSVIGNNLANTSTIGFKK37 pKa = 10.64SSTMQFEE44 pKa = 5.14DD45 pKa = 3.43IFYY48 pKa = 10.69QSVNTGAGIGQVGVGAGVAAIYY70 pKa = 11.02SNFAQGSYY78 pKa = 10.48EE79 pKa = 4.24STSEE83 pKa = 4.03ATDD86 pKa = 3.38VAIGGTGFFMVNNPNTDD103 pKa = 2.65EE104 pKa = 4.1MYY106 pKa = 8.9YY107 pKa = 9.98TRR109 pKa = 11.84AGEE112 pKa = 3.95FRR114 pKa = 11.84FDD116 pKa = 3.23NSGYY120 pKa = 10.78LVDD123 pKa = 3.78TNGYY127 pKa = 8.89RR128 pKa = 11.84VQGWQVKK135 pKa = 9.81DD136 pKa = 3.87GSSSGTVQTTGVSGDD151 pKa = 3.32IQLKK155 pKa = 9.89NFQSPPEE162 pKa = 4.13ATSTVSMLLNLDD174 pKa = 3.79SSSDD178 pKa = 3.74DD179 pKa = 3.94DD180 pKa = 4.28SANAANPFFSLFGNWDD196 pKa = 3.49GTDD199 pKa = 3.37SDD201 pKa = 4.44TPIADD206 pKa = 3.35SRR208 pKa = 11.84YY209 pKa = 9.74AYY211 pKa = 10.69QNTITVYY218 pKa = 10.78DD219 pKa = 4.2EE220 pKa = 4.38NGSAQTLTVYY230 pKa = 10.72CDD232 pKa = 3.53PVKK235 pKa = 10.71DD236 pKa = 3.7ASVVSNSGGNLQWEE250 pKa = 5.19FIVACDD256 pKa = 3.57PSEE259 pKa = 4.43DD260 pKa = 3.3GRR262 pKa = 11.84TIGGQKK268 pKa = 10.1VGSTSAAGLLMTGTLTFNAAGQLTGMSAYY297 pKa = 8.93TLASGASGDD306 pKa = 5.44LKK308 pKa = 11.14DD309 pKa = 4.11LSNWTTADD317 pKa = 4.0LNDD320 pKa = 4.93DD321 pKa = 3.92GLPTFTANFTGEE333 pKa = 4.03ADD335 pKa = 3.36ASATGEE341 pKa = 4.14ANASAIAIDD350 pKa = 4.69FGIHH354 pKa = 7.31DD355 pKa = 4.7SGSPTSWSGSASNASMVGNNPSMLDD380 pKa = 3.15NFTSPKK386 pKa = 9.91ISALTTTSYY395 pKa = 11.29DD396 pKa = 3.4SSSTTISQSQDD407 pKa = 2.54GYY409 pKa = 11.72GPGFLTDD416 pKa = 3.37ISVNRR421 pKa = 11.84DD422 pKa = 2.85GVITGTYY429 pKa = 10.73SNGQVLDD436 pKa = 4.39LYY438 pKa = 11.38VLTLASFTNPYY449 pKa = 9.8GLSRR453 pKa = 11.84EE454 pKa = 3.92GDD456 pKa = 3.55NLFSATRR463 pKa = 11.84EE464 pKa = 3.79SGAAVTGTANTGQLGSISSNTLEE487 pKa = 4.14QSNVDD492 pKa = 3.52TATEE496 pKa = 4.09MVDD499 pKa = 5.24LITTQRR505 pKa = 11.84GFEE508 pKa = 4.22ANSKK512 pKa = 10.68VITTADD518 pKa = 3.25SMLSEE523 pKa = 5.48LIQLKK528 pKa = 10.17RR529 pKa = 3.41
Molecular weight: 55.07 kDa
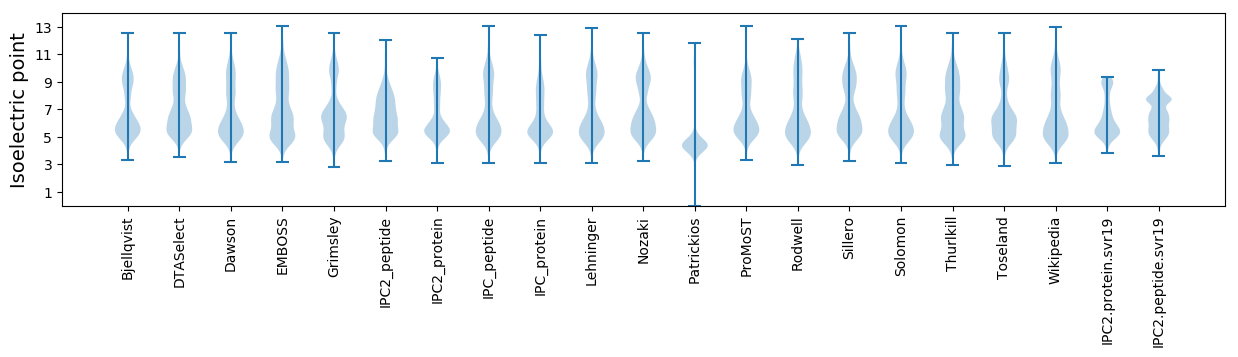
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S7V9I9|S7V9I9_9DELT Acetolactate synthase OS=Desulfovibrio sp. X2 OX=941449 GN=dsx2_2403 PE=4 SV=1
MM1 pKa = 7.66SSRR4 pKa = 11.84LTLVIRR10 pKa = 11.84VDD12 pKa = 4.33FKK14 pKa = 10.19TRR16 pKa = 11.84RR17 pKa = 11.84VLAVFGGRR25 pKa = 11.84RR26 pKa = 11.84SRR28 pKa = 11.84LAPAAPRR35 pKa = 11.84APLPAHH41 pKa = 7.15LLRR44 pKa = 11.84LTTGAATASQGGAQQ58 pKa = 3.48
MM1 pKa = 7.66SSRR4 pKa = 11.84LTLVIRR10 pKa = 11.84VDD12 pKa = 4.33FKK14 pKa = 10.19TRR16 pKa = 11.84RR17 pKa = 11.84VLAVFGGRR25 pKa = 11.84RR26 pKa = 11.84SRR28 pKa = 11.84LAPAAPRR35 pKa = 11.84APLPAHH41 pKa = 7.15LLRR44 pKa = 11.84LTTGAATASQGGAQQ58 pKa = 3.48
Molecular weight: 6.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1149838 |
32 |
1807 |
333.9 |
36.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.984 ± 0.065 | 1.344 ± 0.018 |
5.406 ± 0.028 | 6.493 ± 0.042 |
3.817 ± 0.026 | 8.652 ± 0.035 |
2.027 ± 0.02 | 4.271 ± 0.035 |
3.678 ± 0.039 | 11.096 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.533 ± 0.017 | 2.328 ± 0.021 |
5.227 ± 0.032 | 2.895 ± 0.026 |
7.321 ± 0.044 | 5.338 ± 0.028 |
4.718 ± 0.026 | 7.392 ± 0.033 |
1.185 ± 0.015 | 2.295 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
