
Mameliella alba
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Mameliella
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
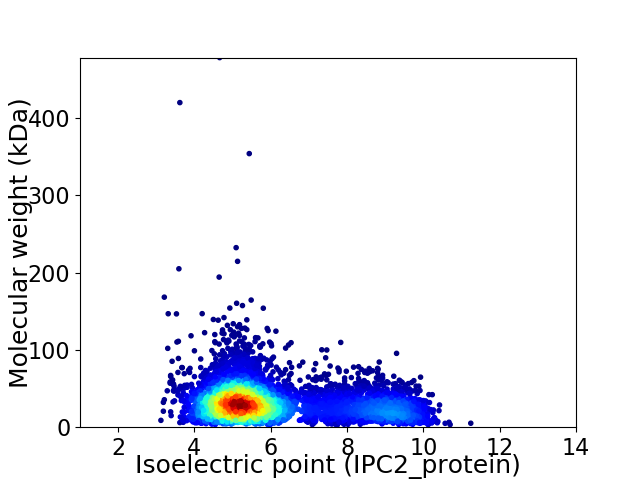
Virtual 2D-PAGE plot for 5714 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B3RIY3|A0A0B3RIY3_9RHOB Uncharacterized protein OS=Mameliella alba OX=561184 GN=GGR29_000948 PE=4 SV=1
MM1 pKa = 7.78CFLCQSLDD9 pKa = 3.68PSIEE13 pKa = 3.87IYY15 pKa = 10.45DD16 pKa = 3.78GHH18 pKa = 7.15GLTGPTAAPSGANGGTTEE36 pKa = 3.84AALPVFTLDD45 pKa = 3.18QVAYY49 pKa = 10.05QLTHH53 pKa = 7.01GYY55 pKa = 7.25WQNGGGDD62 pKa = 2.99WRR64 pKa = 11.84AFDD67 pKa = 3.64VQAGGEE73 pKa = 4.0ITVNIDD79 pKa = 3.09ALDD82 pKa = 4.01AAGQTAALAALEE94 pKa = 4.16AWTAVSGIQFTLTSGTADD112 pKa = 2.79ITFDD116 pKa = 3.63DD117 pKa = 4.57TEE119 pKa = 5.08SGAYY123 pKa = 9.58AYY125 pKa = 10.66SYY127 pKa = 11.52VYY129 pKa = 10.14TSWNEE134 pKa = 3.36IDD136 pKa = 3.35QSFVNVHH143 pKa = 6.59PSWQGYY149 pKa = 8.29GDD151 pKa = 4.26YY152 pKa = 10.93YY153 pKa = 9.0YY154 pKa = 9.58QTYY157 pKa = 8.98IHH159 pKa = 7.24EE160 pKa = 4.73IGHH163 pKa = 6.46ALGLGHH169 pKa = 7.18GGNYY173 pKa = 9.87NGAADD178 pKa = 4.89FGTDD182 pKa = 2.45AHH184 pKa = 6.43YY185 pKa = 11.5ANDD188 pKa = 3.48SWQMSIMSYY197 pKa = 9.67FAQWEE202 pKa = 4.36NPNVDD207 pKa = 3.05ASGMYY212 pKa = 10.37LATPQMADD220 pKa = 2.5ILAIQNLYY228 pKa = 8.15GTPDD232 pKa = 3.32NVHH235 pKa = 6.49TGNTVYY241 pKa = 11.13GDD243 pKa = 3.84NTNLTATGMDD253 pKa = 4.67LAPGRR258 pKa = 11.84AVAIFDD264 pKa = 3.56SSGIDD269 pKa = 3.63VIDD272 pKa = 4.1LGSRR276 pKa = 11.84GHH278 pKa = 6.02DD279 pKa = 2.91QRR281 pKa = 11.84LSLEE285 pKa = 4.24PEE287 pKa = 3.78SWSDD291 pKa = 2.82IDD293 pKa = 5.07GYY295 pKa = 11.56AGNFAIARR303 pKa = 11.84GAVIEE308 pKa = 3.95NAITGSGDD316 pKa = 3.27DD317 pKa = 4.83HH318 pKa = 6.27ITGNAASNDD327 pKa = 3.63IQSGAGADD335 pKa = 3.8TIKK338 pKa = 10.91GGEE341 pKa = 4.22GDD343 pKa = 3.75DD344 pKa = 4.55TIDD347 pKa = 3.52GGAGEE352 pKa = 4.56DD353 pKa = 3.44VVVFEE358 pKa = 4.82GASEE362 pKa = 4.25GFALAWNGALTVTDD376 pKa = 3.95IDD378 pKa = 4.24LTDD381 pKa = 4.28GGDD384 pKa = 3.86DD385 pKa = 3.99GTDD388 pKa = 2.92TLLGVEE394 pKa = 4.55TLSFTDD400 pKa = 4.24GAFGTVQDD408 pKa = 4.43NGTTTLVRR416 pKa = 11.84LYY418 pKa = 9.21QTGSNLTAATLEE430 pKa = 3.85IDD432 pKa = 4.41VDD434 pKa = 4.37DD435 pKa = 3.76THH437 pKa = 6.55EE438 pKa = 4.14WEE440 pKa = 4.8SIARR444 pKa = 11.84DD445 pKa = 4.07FTATGQWAKK454 pKa = 8.25QTNTYY459 pKa = 11.31DD460 pKa = 3.1NGRR463 pKa = 11.84VLEE466 pKa = 4.13IFYY469 pKa = 10.92SDD471 pKa = 3.85GLRR474 pKa = 11.84TSSTMTDD481 pKa = 3.13SADD484 pKa = 3.2VYY486 pKa = 11.37AWDD489 pKa = 4.75SYY491 pKa = 10.41TDD493 pKa = 4.09GYY495 pKa = 10.54DD496 pKa = 3.09ASGARR501 pKa = 11.84IYY503 pKa = 11.19NGITWDD509 pKa = 3.39GGKK512 pKa = 9.98LVEE515 pKa = 4.42THH517 pKa = 6.79FDD519 pKa = 3.32ADD521 pKa = 3.83GARR524 pKa = 11.84TSSLVTDD531 pKa = 4.43GGDD534 pKa = 3.06VYY536 pKa = 11.13GWHH539 pKa = 6.42TIEE542 pKa = 5.53RR543 pKa = 11.84VYY545 pKa = 11.53DD546 pKa = 3.76DD547 pKa = 5.28AGVLTDD553 pKa = 3.6QTNTYY558 pKa = 11.09DD559 pKa = 3.96DD560 pKa = 4.29GRR562 pKa = 11.84VQQIVYY568 pKa = 9.5TEE570 pKa = 4.25GARR573 pKa = 11.84SAIHH577 pKa = 5.49MTDD580 pKa = 3.18AGDD583 pKa = 3.62VASWASYY590 pKa = 10.46SDD592 pKa = 4.57FYY594 pKa = 11.51ATATGARR601 pKa = 11.84TARR604 pKa = 11.84QMTYY608 pKa = 10.9DD609 pKa = 4.2DD610 pKa = 4.04GRR612 pKa = 11.84QVEE615 pKa = 4.13IGFDD619 pKa = 3.64GGRR622 pKa = 11.84MVSHH626 pKa = 7.58LLTDD630 pKa = 4.07GADD633 pKa = 3.48DD634 pKa = 4.5FVWSSIARR642 pKa = 11.84TWDD645 pKa = 3.08AEE647 pKa = 4.2GQLDD651 pKa = 4.35SQTNTYY657 pKa = 11.15DD658 pKa = 3.31DD659 pKa = 4.15GRR661 pKa = 11.84VHH663 pKa = 7.27EE664 pKa = 4.96IDD666 pKa = 3.63YY667 pKa = 11.2ADD669 pKa = 3.81GVRR672 pKa = 11.84SSSVLTDD679 pKa = 3.37TQDD682 pKa = 2.98AYY684 pKa = 10.92AWTSATEE691 pKa = 4.22TYY693 pKa = 9.99DD694 pKa = 3.23ASGALVEE701 pKa = 5.81RR702 pKa = 11.84VTIWDD707 pKa = 4.54DD708 pKa = 3.41GSQDD712 pKa = 3.25VVTYY716 pKa = 10.71GDD718 pKa = 3.9SPVVV722 pKa = 3.24
MM1 pKa = 7.78CFLCQSLDD9 pKa = 3.68PSIEE13 pKa = 3.87IYY15 pKa = 10.45DD16 pKa = 3.78GHH18 pKa = 7.15GLTGPTAAPSGANGGTTEE36 pKa = 3.84AALPVFTLDD45 pKa = 3.18QVAYY49 pKa = 10.05QLTHH53 pKa = 7.01GYY55 pKa = 7.25WQNGGGDD62 pKa = 2.99WRR64 pKa = 11.84AFDD67 pKa = 3.64VQAGGEE73 pKa = 4.0ITVNIDD79 pKa = 3.09ALDD82 pKa = 4.01AAGQTAALAALEE94 pKa = 4.16AWTAVSGIQFTLTSGTADD112 pKa = 2.79ITFDD116 pKa = 3.63DD117 pKa = 4.57TEE119 pKa = 5.08SGAYY123 pKa = 9.58AYY125 pKa = 10.66SYY127 pKa = 11.52VYY129 pKa = 10.14TSWNEE134 pKa = 3.36IDD136 pKa = 3.35QSFVNVHH143 pKa = 6.59PSWQGYY149 pKa = 8.29GDD151 pKa = 4.26YY152 pKa = 10.93YY153 pKa = 9.0YY154 pKa = 9.58QTYY157 pKa = 8.98IHH159 pKa = 7.24EE160 pKa = 4.73IGHH163 pKa = 6.46ALGLGHH169 pKa = 7.18GGNYY173 pKa = 9.87NGAADD178 pKa = 4.89FGTDD182 pKa = 2.45AHH184 pKa = 6.43YY185 pKa = 11.5ANDD188 pKa = 3.48SWQMSIMSYY197 pKa = 9.67FAQWEE202 pKa = 4.36NPNVDD207 pKa = 3.05ASGMYY212 pKa = 10.37LATPQMADD220 pKa = 2.5ILAIQNLYY228 pKa = 8.15GTPDD232 pKa = 3.32NVHH235 pKa = 6.49TGNTVYY241 pKa = 11.13GDD243 pKa = 3.84NTNLTATGMDD253 pKa = 4.67LAPGRR258 pKa = 11.84AVAIFDD264 pKa = 3.56SSGIDD269 pKa = 3.63VIDD272 pKa = 4.1LGSRR276 pKa = 11.84GHH278 pKa = 6.02DD279 pKa = 2.91QRR281 pKa = 11.84LSLEE285 pKa = 4.24PEE287 pKa = 3.78SWSDD291 pKa = 2.82IDD293 pKa = 5.07GYY295 pKa = 11.56AGNFAIARR303 pKa = 11.84GAVIEE308 pKa = 3.95NAITGSGDD316 pKa = 3.27DD317 pKa = 4.83HH318 pKa = 6.27ITGNAASNDD327 pKa = 3.63IQSGAGADD335 pKa = 3.8TIKK338 pKa = 10.91GGEE341 pKa = 4.22GDD343 pKa = 3.75DD344 pKa = 4.55TIDD347 pKa = 3.52GGAGEE352 pKa = 4.56DD353 pKa = 3.44VVVFEE358 pKa = 4.82GASEE362 pKa = 4.25GFALAWNGALTVTDD376 pKa = 3.95IDD378 pKa = 4.24LTDD381 pKa = 4.28GGDD384 pKa = 3.86DD385 pKa = 3.99GTDD388 pKa = 2.92TLLGVEE394 pKa = 4.55TLSFTDD400 pKa = 4.24GAFGTVQDD408 pKa = 4.43NGTTTLVRR416 pKa = 11.84LYY418 pKa = 9.21QTGSNLTAATLEE430 pKa = 3.85IDD432 pKa = 4.41VDD434 pKa = 4.37DD435 pKa = 3.76THH437 pKa = 6.55EE438 pKa = 4.14WEE440 pKa = 4.8SIARR444 pKa = 11.84DD445 pKa = 4.07FTATGQWAKK454 pKa = 8.25QTNTYY459 pKa = 11.31DD460 pKa = 3.1NGRR463 pKa = 11.84VLEE466 pKa = 4.13IFYY469 pKa = 10.92SDD471 pKa = 3.85GLRR474 pKa = 11.84TSSTMTDD481 pKa = 3.13SADD484 pKa = 3.2VYY486 pKa = 11.37AWDD489 pKa = 4.75SYY491 pKa = 10.41TDD493 pKa = 4.09GYY495 pKa = 10.54DD496 pKa = 3.09ASGARR501 pKa = 11.84IYY503 pKa = 11.19NGITWDD509 pKa = 3.39GGKK512 pKa = 9.98LVEE515 pKa = 4.42THH517 pKa = 6.79FDD519 pKa = 3.32ADD521 pKa = 3.83GARR524 pKa = 11.84TSSLVTDD531 pKa = 4.43GGDD534 pKa = 3.06VYY536 pKa = 11.13GWHH539 pKa = 6.42TIEE542 pKa = 5.53RR543 pKa = 11.84VYY545 pKa = 11.53DD546 pKa = 3.76DD547 pKa = 5.28AGVLTDD553 pKa = 3.6QTNTYY558 pKa = 11.09DD559 pKa = 3.96DD560 pKa = 4.29GRR562 pKa = 11.84VQQIVYY568 pKa = 9.5TEE570 pKa = 4.25GARR573 pKa = 11.84SAIHH577 pKa = 5.49MTDD580 pKa = 3.18AGDD583 pKa = 3.62VASWASYY590 pKa = 10.46SDD592 pKa = 4.57FYY594 pKa = 11.51ATATGARR601 pKa = 11.84TARR604 pKa = 11.84QMTYY608 pKa = 10.9DD609 pKa = 4.2DD610 pKa = 4.04GRR612 pKa = 11.84QVEE615 pKa = 4.13IGFDD619 pKa = 3.64GGRR622 pKa = 11.84MVSHH626 pKa = 7.58LLTDD630 pKa = 4.07GADD633 pKa = 3.48DD634 pKa = 4.5FVWSSIARR642 pKa = 11.84TWDD645 pKa = 3.08AEE647 pKa = 4.2GQLDD651 pKa = 4.35SQTNTYY657 pKa = 11.15DD658 pKa = 3.31DD659 pKa = 4.15GRR661 pKa = 11.84VHH663 pKa = 7.27EE664 pKa = 4.96IDD666 pKa = 3.63YY667 pKa = 11.2ADD669 pKa = 3.81GVRR672 pKa = 11.84SSSVLTDD679 pKa = 3.37TQDD682 pKa = 2.98AYY684 pKa = 10.92AWTSATEE691 pKa = 4.22TYY693 pKa = 9.99DD694 pKa = 3.23ASGALVEE701 pKa = 5.81RR702 pKa = 11.84VTIWDD707 pKa = 4.54DD708 pKa = 3.41GSQDD712 pKa = 3.25VVTYY716 pKa = 10.71GDD718 pKa = 3.9SPVVV722 pKa = 3.24
Molecular weight: 77.07 kDa
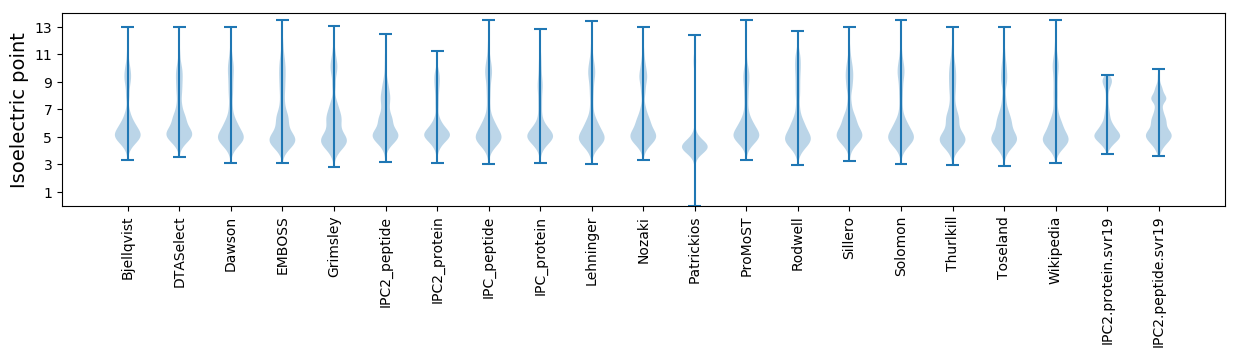
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B3RZK6|A0A0B3RZK6_9RHOB 8-oxo-dGTP diphosphatase OS=Mameliella alba OX=561184 GN=mutT PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1739916 |
29 |
4497 |
304.5 |
33.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.318 ± 0.045 | 0.946 ± 0.012 |
6.257 ± 0.035 | 6.005 ± 0.028 |
3.618 ± 0.019 | 8.972 ± 0.036 |
2.068 ± 0.016 | 4.814 ± 0.024 |
2.878 ± 0.026 | 10.104 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.747 ± 0.018 | 2.433 ± 0.02 |
5.308 ± 0.029 | 3.163 ± 0.015 |
6.993 ± 0.034 | 4.958 ± 0.021 |
5.466 ± 0.027 | 7.286 ± 0.028 |
1.453 ± 0.013 | 2.213 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
