
Blastococcus endophyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
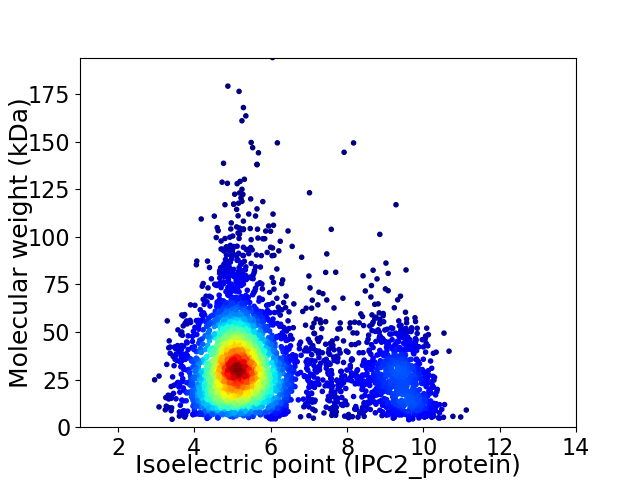
Virtual 2D-PAGE plot for 4606 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8W336|A0A1H8W336_9ACTN DNA-binding transcriptional regulator MarR family OS=Blastococcus endophyticus OX=673521 GN=SAMN05660991_03983 PE=4 SV=1
MM1 pKa = 7.4TPLGSSGRR9 pKa = 11.84RR10 pKa = 11.84VRR12 pKa = 11.84ATTAGIAVTALALTACGGGDD32 pKa = 4.29DD33 pKa = 5.31DD34 pKa = 5.15NGSGGSGGSADD45 pKa = 3.79ALIVGTTDD53 pKa = 3.92KK54 pKa = 10.38ITTIDD59 pKa = 3.49PAGSYY64 pKa = 11.17DD65 pKa = 3.54NGSFAVMNQVYY76 pKa = 9.38PFLLNTPLGSPEE88 pKa = 4.13VEE90 pKa = 3.58PDD92 pKa = 2.97IAEE95 pKa = 4.42SADD98 pKa = 3.54FTSPTQYY105 pKa = 10.7TVTLKK110 pKa = 10.81PGLTFANGNEE120 pKa = 4.04LTASDD125 pKa = 4.03VKK127 pKa = 10.85FSFDD131 pKa = 3.4RR132 pKa = 11.84QVAIADD138 pKa = 3.83EE139 pKa = 4.69NGPSSLLANLEE150 pKa = 4.4SVEE153 pKa = 4.61APDD156 pKa = 3.59DD157 pKa = 3.75TTVVFNLTNPDD168 pKa = 3.74DD169 pKa = 3.76QTFPQVLSSPVGPIVDD185 pKa = 3.98EE186 pKa = 4.58EE187 pKa = 4.61VFSADD192 pKa = 3.86SLTPDD197 pKa = 3.9DD198 pKa = 5.53EE199 pKa = 4.54IVAGNAFAGPYY210 pKa = 10.1VITSYY215 pKa = 10.76EE216 pKa = 4.03INDD219 pKa = 4.67LIAYY223 pKa = 7.35EE224 pKa = 4.76ANPDD228 pKa = 3.66YY229 pKa = 10.91QGLLGAPKK237 pKa = 9.67TDD239 pKa = 4.01TINVRR244 pKa = 11.84YY245 pKa = 9.04YY246 pKa = 11.41AEE248 pKa = 4.26ASNLKK253 pKa = 10.33LDD255 pKa = 3.79IEE257 pKa = 4.46EE258 pKa = 4.47GAVDD262 pKa = 3.24VAYY265 pKa = 10.47RR266 pKa = 11.84SLSATDD272 pKa = 3.87VEE274 pKa = 4.61DD275 pKa = 4.59LRR277 pKa = 11.84GNDD280 pKa = 3.46DD281 pKa = 3.71VEE283 pKa = 4.74VVDD286 pKa = 5.11GPGGEE291 pKa = 3.55IRR293 pKa = 11.84YY294 pKa = 9.33IVFNFNTQPYY304 pKa = 9.14GATTAEE310 pKa = 4.31ADD312 pKa = 3.66PAKK315 pKa = 10.67ALAVRR320 pKa = 11.84QAMAHH325 pKa = 6.32LIDD328 pKa = 4.28RR329 pKa = 11.84EE330 pKa = 4.02EE331 pKa = 4.27LAEE334 pKa = 3.89QVYY337 pKa = 10.47KK338 pKa = 10.16GTYY341 pKa = 7.52TPLYY345 pKa = 10.12SYY347 pKa = 10.96VADD350 pKa = 4.27GLTGATDD357 pKa = 3.71SLRR360 pKa = 11.84GLYY363 pKa = 10.57GDD365 pKa = 4.08GQGGPDD371 pKa = 3.86ADD373 pKa = 3.79AAAQVLEE380 pKa = 4.23AAGVEE385 pKa = 4.58TPVQLSLQYY394 pKa = 11.43SNDD397 pKa = 3.17HH398 pKa = 6.35YY399 pKa = 11.31GPSSGDD405 pKa = 3.07EE406 pKa = 3.82YY407 pKa = 11.92ALIKK411 pKa = 10.68DD412 pKa = 3.77QLEE415 pKa = 4.36STGLFTVDD423 pKa = 3.92LQTTEE428 pKa = 3.39WTQYY432 pKa = 10.48SEE434 pKa = 4.06QRR436 pKa = 11.84TADD439 pKa = 3.6TYY441 pKa = 10.9PAYY444 pKa = 10.2QLGWFPDD451 pKa = 3.67YY452 pKa = 11.48SDD454 pKa = 4.27ADD456 pKa = 3.57NYY458 pKa = 10.67LAPFFLTEE466 pKa = 4.09NFLSNHH472 pKa = 6.16YY473 pKa = 10.76DD474 pKa = 3.48NQQVNDD480 pKa = 5.53LILQQLSTPDD490 pKa = 3.13EE491 pKa = 4.24AARR494 pKa = 11.84TALIEE499 pKa = 4.06QIQDD503 pKa = 3.38LVAADD508 pKa = 5.24LSTLPYY514 pKa = 10.43LQGAQIAVVRR524 pKa = 11.84TGVTGAEE531 pKa = 4.21DD532 pKa = 3.46TLDD535 pKa = 3.3ASFKK539 pKa = 10.23FRR541 pKa = 11.84YY542 pKa = 9.05GALEE546 pKa = 3.81RR547 pKa = 11.84AA548 pKa = 4.11
MM1 pKa = 7.4TPLGSSGRR9 pKa = 11.84RR10 pKa = 11.84VRR12 pKa = 11.84ATTAGIAVTALALTACGGGDD32 pKa = 4.29DD33 pKa = 5.31DD34 pKa = 5.15NGSGGSGGSADD45 pKa = 3.79ALIVGTTDD53 pKa = 3.92KK54 pKa = 10.38ITTIDD59 pKa = 3.49PAGSYY64 pKa = 11.17DD65 pKa = 3.54NGSFAVMNQVYY76 pKa = 9.38PFLLNTPLGSPEE88 pKa = 4.13VEE90 pKa = 3.58PDD92 pKa = 2.97IAEE95 pKa = 4.42SADD98 pKa = 3.54FTSPTQYY105 pKa = 10.7TVTLKK110 pKa = 10.81PGLTFANGNEE120 pKa = 4.04LTASDD125 pKa = 4.03VKK127 pKa = 10.85FSFDD131 pKa = 3.4RR132 pKa = 11.84QVAIADD138 pKa = 3.83EE139 pKa = 4.69NGPSSLLANLEE150 pKa = 4.4SVEE153 pKa = 4.61APDD156 pKa = 3.59DD157 pKa = 3.75TTVVFNLTNPDD168 pKa = 3.74DD169 pKa = 3.76QTFPQVLSSPVGPIVDD185 pKa = 3.98EE186 pKa = 4.58EE187 pKa = 4.61VFSADD192 pKa = 3.86SLTPDD197 pKa = 3.9DD198 pKa = 5.53EE199 pKa = 4.54IVAGNAFAGPYY210 pKa = 10.1VITSYY215 pKa = 10.76EE216 pKa = 4.03INDD219 pKa = 4.67LIAYY223 pKa = 7.35EE224 pKa = 4.76ANPDD228 pKa = 3.66YY229 pKa = 10.91QGLLGAPKK237 pKa = 9.67TDD239 pKa = 4.01TINVRR244 pKa = 11.84YY245 pKa = 9.04YY246 pKa = 11.41AEE248 pKa = 4.26ASNLKK253 pKa = 10.33LDD255 pKa = 3.79IEE257 pKa = 4.46EE258 pKa = 4.47GAVDD262 pKa = 3.24VAYY265 pKa = 10.47RR266 pKa = 11.84SLSATDD272 pKa = 3.87VEE274 pKa = 4.61DD275 pKa = 4.59LRR277 pKa = 11.84GNDD280 pKa = 3.46DD281 pKa = 3.71VEE283 pKa = 4.74VVDD286 pKa = 5.11GPGGEE291 pKa = 3.55IRR293 pKa = 11.84YY294 pKa = 9.33IVFNFNTQPYY304 pKa = 9.14GATTAEE310 pKa = 4.31ADD312 pKa = 3.66PAKK315 pKa = 10.67ALAVRR320 pKa = 11.84QAMAHH325 pKa = 6.32LIDD328 pKa = 4.28RR329 pKa = 11.84EE330 pKa = 4.02EE331 pKa = 4.27LAEE334 pKa = 3.89QVYY337 pKa = 10.47KK338 pKa = 10.16GTYY341 pKa = 7.52TPLYY345 pKa = 10.12SYY347 pKa = 10.96VADD350 pKa = 4.27GLTGATDD357 pKa = 3.71SLRR360 pKa = 11.84GLYY363 pKa = 10.57GDD365 pKa = 4.08GQGGPDD371 pKa = 3.86ADD373 pKa = 3.79AAAQVLEE380 pKa = 4.23AAGVEE385 pKa = 4.58TPVQLSLQYY394 pKa = 11.43SNDD397 pKa = 3.17HH398 pKa = 6.35YY399 pKa = 11.31GPSSGDD405 pKa = 3.07EE406 pKa = 3.82YY407 pKa = 11.92ALIKK411 pKa = 10.68DD412 pKa = 3.77QLEE415 pKa = 4.36STGLFTVDD423 pKa = 3.92LQTTEE428 pKa = 3.39WTQYY432 pKa = 10.48SEE434 pKa = 4.06QRR436 pKa = 11.84TADD439 pKa = 3.6TYY441 pKa = 10.9PAYY444 pKa = 10.2QLGWFPDD451 pKa = 3.67YY452 pKa = 11.48SDD454 pKa = 4.27ADD456 pKa = 3.57NYY458 pKa = 10.67LAPFFLTEE466 pKa = 4.09NFLSNHH472 pKa = 6.16YY473 pKa = 10.76DD474 pKa = 3.48NQQVNDD480 pKa = 5.53LILQQLSTPDD490 pKa = 3.13EE491 pKa = 4.24AARR494 pKa = 11.84TALIEE499 pKa = 4.06QIQDD503 pKa = 3.38LVAADD508 pKa = 5.24LSTLPYY514 pKa = 10.43LQGAQIAVVRR524 pKa = 11.84TGVTGAEE531 pKa = 4.21DD532 pKa = 3.46TLDD535 pKa = 3.3ASFKK539 pKa = 10.23FRR541 pKa = 11.84YY542 pKa = 9.05GALEE546 pKa = 3.81RR547 pKa = 11.84AA548 pKa = 4.11
Molecular weight: 58.45 kDa
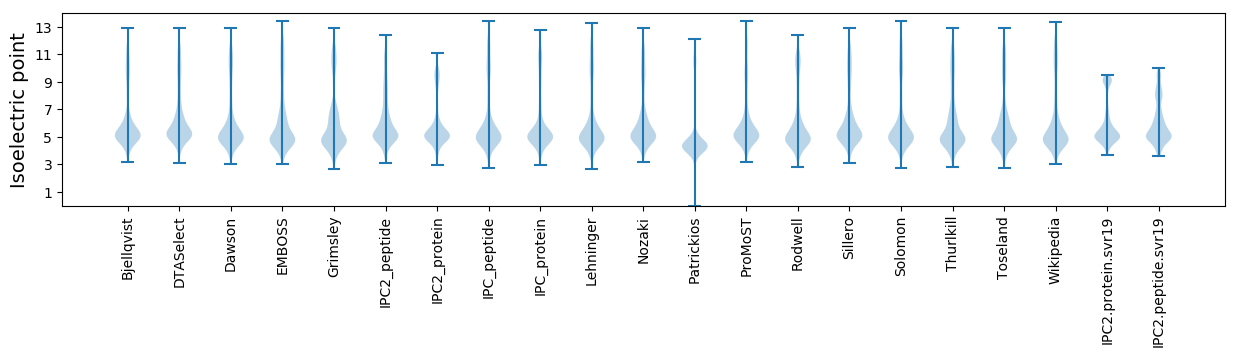
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8WJV0|A0A1H8WJV0_9ACTN Methyltransferase domain-containing protein OS=Blastococcus endophyticus OX=673521 GN=SAMN05660991_04484 PE=4 SV=1
MM1 pKa = 7.12LTVALAWTAVLASVVAAALFGQTARR26 pKa = 11.84GGAPRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SVARR39 pKa = 11.84TRR41 pKa = 11.84AGLAVAMSGPWLVLVLGVAAGAVTGAWLAAAAGTVAGVVAVATAGLVLVPRR92 pKa = 4.98
MM1 pKa = 7.12LTVALAWTAVLASVVAAALFGQTARR26 pKa = 11.84GGAPRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SVARR39 pKa = 11.84TRR41 pKa = 11.84AGLAVAMSGPWLVLVLGVAAGAVTGAWLAAAAGTVAGVVAVATAGLVLVPRR92 pKa = 4.98
Molecular weight: 9.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1481554 |
39 |
1827 |
321.7 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.708 ± 0.057 | 0.705 ± 0.011 |
6.297 ± 0.029 | 5.64 ± 0.031 |
2.598 ± 0.021 | 9.836 ± 0.032 |
1.971 ± 0.016 | 2.818 ± 0.025 |
1.275 ± 0.022 | 10.592 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.636 ± 0.015 | 1.411 ± 0.016 |
6.35 ± 0.038 | 2.521 ± 0.02 |
8.198 ± 0.037 | 4.689 ± 0.025 |
5.831 ± 0.025 | 9.699 ± 0.04 |
1.462 ± 0.015 | 1.762 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
