
Salmonella phage SEN4
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Senquatrovirus; Salmonella virus SEN4
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
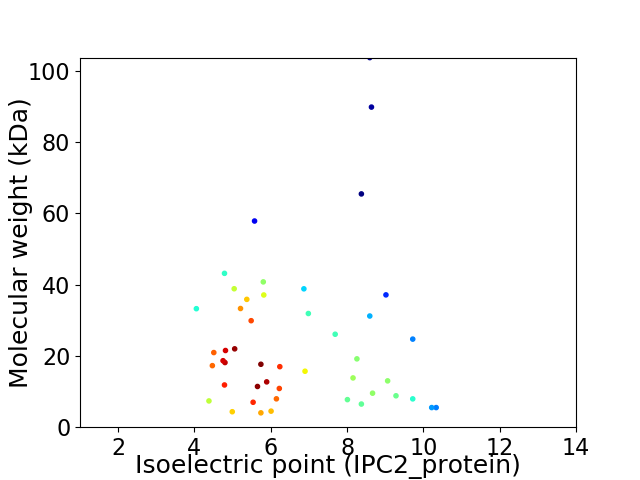
Virtual 2D-PAGE plot for 47 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
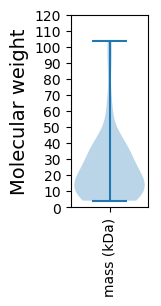
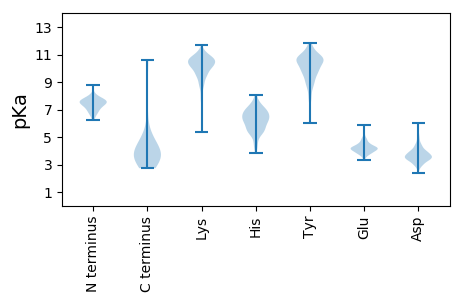
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0M4R507|A0A0M4R507_9CAUD Late control D protein OS=Salmonella phage SEN4 OX=1647465 GN=SEN4_28 PE=4 SV=1
MM1 pKa = 7.64SGTIDD6 pKa = 4.84LSQLPPPVVVEE17 pKa = 4.06PLDD20 pKa = 3.89FEE22 pKa = 4.7TLFAQRR28 pKa = 11.84KK29 pKa = 6.95AAFIAMYY36 pKa = 10.23PEE38 pKa = 4.92DD39 pKa = 4.31EE40 pKa = 4.28QEE42 pKa = 4.23EE43 pKa = 4.27IARR46 pKa = 11.84TLEE49 pKa = 4.3LEE51 pKa = 4.2SEE53 pKa = 4.87PITMLLEE60 pKa = 4.14EE61 pKa = 4.03NCYY64 pKa = 10.44RR65 pKa = 11.84EE66 pKa = 4.01LLLRR70 pKa = 11.84QRR72 pKa = 11.84VNEE75 pKa = 3.92AARR78 pKa = 11.84AVMLAYY84 pKa = 9.8STDD87 pKa = 3.45SDD89 pKa = 4.56LDD91 pKa = 3.66NLAVNFNVEE100 pKa = 3.66RR101 pKa = 11.84LTIQEE106 pKa = 4.41EE107 pKa = 4.42DD108 pKa = 4.01DD109 pKa = 4.2SVTPPIEE116 pKa = 4.01AVVEE120 pKa = 4.06SDD122 pKa = 2.72ADD124 pKa = 3.56LRR126 pKa = 11.84TRR128 pKa = 11.84TQQAFEE134 pKa = 4.23GLSVAGPTAAYY145 pKa = 8.17EE146 pKa = 4.04FWGRR150 pKa = 11.84SADD153 pKa = 3.49GRR155 pKa = 11.84VADD158 pKa = 4.7ISAVSPTPACVTISVLSRR176 pKa = 11.84EE177 pKa = 4.37GDD179 pKa = 3.47GTASDD184 pKa = 4.66DD185 pKa = 3.98LLSVVAAALNDD196 pKa = 3.71EE197 pKa = 4.58EE198 pKa = 4.51VRR200 pKa = 11.84PVADD204 pKa = 3.01RR205 pKa = 11.84VTVQSADD212 pKa = 3.06IVPYY216 pKa = 10.65QIDD219 pKa = 3.74ATLYY223 pKa = 9.62IYY225 pKa = 9.19PGPEE229 pKa = 3.79AEE231 pKa = 4.54PVRR234 pKa = 11.84QASEE238 pKa = 4.05QQLQAYY244 pKa = 6.87ITAQNRR250 pKa = 11.84LGRR253 pKa = 11.84DD254 pKa = 2.99IRR256 pKa = 11.84ISAIYY261 pKa = 9.65AALHH265 pKa = 5.3VEE267 pKa = 4.81GVQRR271 pKa = 11.84VEE273 pKa = 3.98LAQPVADD280 pKa = 4.37IVLSDD285 pKa = 3.81YY286 pKa = 10.63QASHH290 pKa = 5.6CTEE293 pKa = 3.71YY294 pKa = 10.62TITVGGYY301 pKa = 10.39DD302 pKa = 3.42EE303 pKa = 4.8
MM1 pKa = 7.64SGTIDD6 pKa = 4.84LSQLPPPVVVEE17 pKa = 4.06PLDD20 pKa = 3.89FEE22 pKa = 4.7TLFAQRR28 pKa = 11.84KK29 pKa = 6.95AAFIAMYY36 pKa = 10.23PEE38 pKa = 4.92DD39 pKa = 4.31EE40 pKa = 4.28QEE42 pKa = 4.23EE43 pKa = 4.27IARR46 pKa = 11.84TLEE49 pKa = 4.3LEE51 pKa = 4.2SEE53 pKa = 4.87PITMLLEE60 pKa = 4.14EE61 pKa = 4.03NCYY64 pKa = 10.44RR65 pKa = 11.84EE66 pKa = 4.01LLLRR70 pKa = 11.84QRR72 pKa = 11.84VNEE75 pKa = 3.92AARR78 pKa = 11.84AVMLAYY84 pKa = 9.8STDD87 pKa = 3.45SDD89 pKa = 4.56LDD91 pKa = 3.66NLAVNFNVEE100 pKa = 3.66RR101 pKa = 11.84LTIQEE106 pKa = 4.41EE107 pKa = 4.42DD108 pKa = 4.01DD109 pKa = 4.2SVTPPIEE116 pKa = 4.01AVVEE120 pKa = 4.06SDD122 pKa = 2.72ADD124 pKa = 3.56LRR126 pKa = 11.84TRR128 pKa = 11.84TQQAFEE134 pKa = 4.23GLSVAGPTAAYY145 pKa = 8.17EE146 pKa = 4.04FWGRR150 pKa = 11.84SADD153 pKa = 3.49GRR155 pKa = 11.84VADD158 pKa = 4.7ISAVSPTPACVTISVLSRR176 pKa = 11.84EE177 pKa = 4.37GDD179 pKa = 3.47GTASDD184 pKa = 4.66DD185 pKa = 3.98LLSVVAAALNDD196 pKa = 3.71EE197 pKa = 4.58EE198 pKa = 4.51VRR200 pKa = 11.84PVADD204 pKa = 3.01RR205 pKa = 11.84VTVQSADD212 pKa = 3.06IVPYY216 pKa = 10.65QIDD219 pKa = 3.74ATLYY223 pKa = 9.62IYY225 pKa = 9.19PGPEE229 pKa = 3.79AEE231 pKa = 4.54PVRR234 pKa = 11.84QASEE238 pKa = 4.05QQLQAYY244 pKa = 6.87ITAQNRR250 pKa = 11.84LGRR253 pKa = 11.84DD254 pKa = 2.99IRR256 pKa = 11.84ISAIYY261 pKa = 9.65AALHH265 pKa = 5.3VEE267 pKa = 4.81GVQRR271 pKa = 11.84VEE273 pKa = 3.98LAQPVADD280 pKa = 4.37IVLSDD285 pKa = 3.81YY286 pKa = 10.63QASHH290 pKa = 5.6CTEE293 pKa = 3.71YY294 pKa = 10.62TITVGGYY301 pKa = 10.39DD302 pKa = 3.42EE303 pKa = 4.8
Molecular weight: 33.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M5M1H9|A0A0M5M1H9_9CAUD Integrase OS=Salmonella phage SEN4 OX=1647465 GN=SEN4_31 PE=3 SV=1
MM1 pKa = 7.74TGQQTDD7 pKa = 3.79PLFQQLDD14 pKa = 3.38DD15 pKa = 3.49WLARR19 pKa = 11.84VAAQLSPGHH28 pKa = 5.97RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 8.87LTRR34 pKa = 11.84DD35 pKa = 3.19VAIGLRR41 pKa = 11.84KK42 pKa = 9.46RR43 pKa = 11.84QQKK46 pKa = 10.28RR47 pKa = 11.84IASQKK52 pKa = 8.7NPSGEE57 pKa = 4.15SYY59 pKa = 9.16QARR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.45ILRR68 pKa = 11.84TQGGIKK74 pKa = 9.63FIWNDD79 pKa = 3.07EE80 pKa = 3.84ARR82 pKa = 11.84EE83 pKa = 3.95LRR85 pKa = 11.84NWRR88 pKa = 11.84TTGRR92 pKa = 11.84GEE94 pKa = 3.67HH95 pKa = 6.29RR96 pKa = 11.84AITGYY101 pKa = 10.81DD102 pKa = 3.18VDD104 pKa = 4.59RR105 pKa = 11.84GALRR109 pKa = 11.84TFYY112 pKa = 10.87KK113 pKa = 10.27RR114 pKa = 11.84DD115 pKa = 2.95IQRR118 pKa = 11.84YY119 pKa = 8.88IEE121 pKa = 4.19INLNQSKK128 pKa = 9.4QNRR131 pKa = 11.84TRR133 pKa = 11.84KK134 pKa = 10.36DD135 pKa = 3.07PMFRR139 pKa = 11.84KK140 pKa = 9.77LRR142 pKa = 11.84TARR145 pKa = 11.84FLKK148 pKa = 10.65AYY150 pKa = 6.05GTSGIAVVGFQGHH163 pKa = 4.6TAEE166 pKa = 4.51IASVHH171 pKa = 5.79QYY173 pKa = 11.53GEE175 pKa = 3.9VDD177 pKa = 3.46NVVPGARR184 pKa = 11.84ARR186 pKa = 11.84YY187 pKa = 7.83PVRR190 pKa = 11.84EE191 pKa = 4.09LLGMTEE197 pKa = 5.85GDD199 pKa = 4.56LDD201 pKa = 3.8WLADD205 pKa = 3.66TVVAFMQEE213 pKa = 3.51II214 pKa = 3.73
MM1 pKa = 7.74TGQQTDD7 pKa = 3.79PLFQQLDD14 pKa = 3.38DD15 pKa = 3.49WLARR19 pKa = 11.84VAAQLSPGHH28 pKa = 5.97RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 8.87LTRR34 pKa = 11.84DD35 pKa = 3.19VAIGLRR41 pKa = 11.84KK42 pKa = 9.46RR43 pKa = 11.84QQKK46 pKa = 10.28RR47 pKa = 11.84IASQKK52 pKa = 8.7NPSGEE57 pKa = 4.15SYY59 pKa = 9.16QARR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.45ILRR68 pKa = 11.84TQGGIKK74 pKa = 9.63FIWNDD79 pKa = 3.07EE80 pKa = 3.84ARR82 pKa = 11.84EE83 pKa = 3.95LRR85 pKa = 11.84NWRR88 pKa = 11.84TTGRR92 pKa = 11.84GEE94 pKa = 3.67HH95 pKa = 6.29RR96 pKa = 11.84AITGYY101 pKa = 10.81DD102 pKa = 3.18VDD104 pKa = 4.59RR105 pKa = 11.84GALRR109 pKa = 11.84TFYY112 pKa = 10.87KK113 pKa = 10.27RR114 pKa = 11.84DD115 pKa = 2.95IQRR118 pKa = 11.84YY119 pKa = 8.88IEE121 pKa = 4.19INLNQSKK128 pKa = 9.4QNRR131 pKa = 11.84TRR133 pKa = 11.84KK134 pKa = 10.36DD135 pKa = 3.07PMFRR139 pKa = 11.84KK140 pKa = 9.77LRR142 pKa = 11.84TARR145 pKa = 11.84FLKK148 pKa = 10.65AYY150 pKa = 6.05GTSGIAVVGFQGHH163 pKa = 4.6TAEE166 pKa = 4.51IASVHH171 pKa = 5.79QYY173 pKa = 11.53GEE175 pKa = 3.9VDD177 pKa = 3.46NVVPGARR184 pKa = 11.84ARR186 pKa = 11.84YY187 pKa = 7.83PVRR190 pKa = 11.84EE191 pKa = 4.09LLGMTEE197 pKa = 5.85GDD199 pKa = 4.56LDD201 pKa = 3.8WLADD205 pKa = 3.66TVVAFMQEE213 pKa = 3.51II214 pKa = 3.73
Molecular weight: 24.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10358 |
38 |
974 |
220.4 |
24.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.34 ± 0.59 | 1.197 ± 0.191 |
6.15 ± 0.295 | 5.793 ± 0.31 |
3.437 ± 0.318 | 7.289 ± 0.296 |
1.757 ± 0.189 | 5.252 ± 0.209 |
5.223 ± 0.326 | 8.37 ± 0.314 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.159 | 3.92 ± 0.199 |
4.518 ± 0.207 | 4.364 ± 0.381 |
6.343 ± 0.507 | 6.179 ± 0.323 |
6.246 ± 0.401 | 6.488 ± 0.226 |
1.574 ± 0.163 | 3.196 ± 0.241 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
