
Firmicutes bacterium CAG:24
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
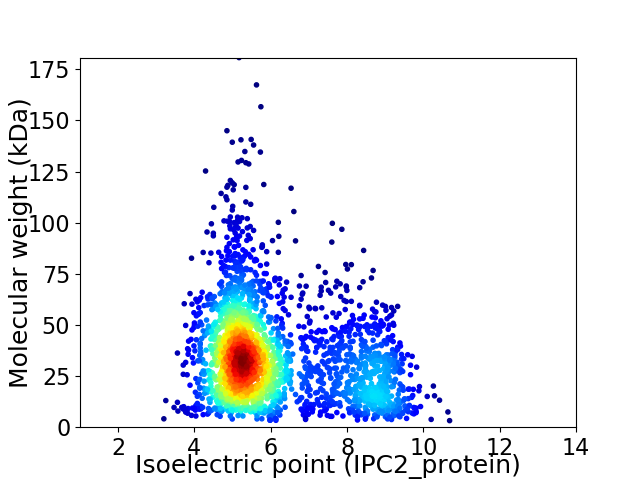
Virtual 2D-PAGE plot for 2518 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
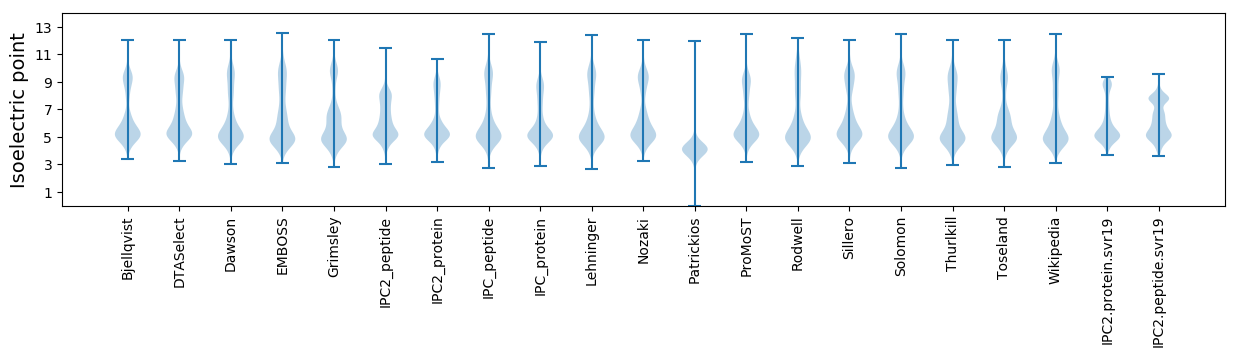
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5H0B2|R5H0B2_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:24 OX=1263012 GN=BN555_00317 PE=4 SV=1
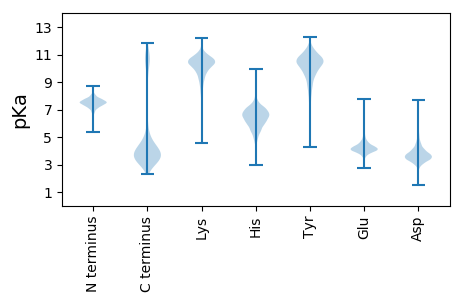
MM1 pKa = 7.5SMKK4 pKa = 10.55KK5 pKa = 10.16LGSILLSTAMVMSLAACGSQNTSAPAATTDD35 pKa = 3.54DD36 pKa = 3.86SATKK40 pKa = 10.27EE41 pKa = 4.07EE42 pKa = 4.23AVAPAEE48 pKa = 4.28SAVAADD54 pKa = 3.94TDD56 pKa = 4.11VSGSTLEE63 pKa = 4.09VAVTYY68 pKa = 10.21TGDD71 pKa = 3.31QATTFQGLVDD81 pKa = 4.02KK82 pKa = 11.21FEE84 pKa = 4.21EE85 pKa = 4.88EE86 pKa = 4.25YY87 pKa = 11.23GVTVNIAEE95 pKa = 4.29YY96 pKa = 11.04GDD98 pKa = 5.13DD99 pKa = 4.15YY100 pKa = 11.87EE101 pKa = 4.53NTLKK105 pKa = 9.92TRR107 pKa = 11.84MASNEE112 pKa = 4.09LPDD115 pKa = 3.65VFQTHH120 pKa = 5.39GWSLLRR126 pKa = 11.84YY127 pKa = 9.47KK128 pKa = 10.27EE129 pKa = 4.11YY130 pKa = 11.16LMNLQDD136 pKa = 4.08QPWVSDD142 pKa = 3.66YY143 pKa = 11.59DD144 pKa = 3.88EE145 pKa = 4.79SALGVIQDD153 pKa = 4.5DD154 pKa = 4.02DD155 pKa = 4.0GAIYY159 pKa = 11.11VLMISEE165 pKa = 5.79LINGTLVNLDD175 pKa = 3.44ACDD178 pKa = 3.42AAGVDD183 pKa = 4.65PYY185 pKa = 11.29AIHH188 pKa = 6.29TWDD191 pKa = 5.19DD192 pKa = 3.41FTEE195 pKa = 4.38ACAKK199 pKa = 10.17IKK201 pKa = 10.12TSGLTPLNVVSNPGLLANFAGTFVSYY227 pKa = 10.01EE228 pKa = 3.92GEE230 pKa = 4.11MFEE233 pKa = 6.1DD234 pKa = 3.67SAAMLDD240 pKa = 5.56GSFDD244 pKa = 3.31WQDD247 pKa = 3.44YY248 pKa = 10.38KK249 pKa = 11.61DD250 pKa = 3.91SLVKK254 pKa = 10.65YY255 pKa = 6.7MANWIDD261 pKa = 2.94SGYY264 pKa = 10.79YY265 pKa = 9.74YY266 pKa = 10.97DD267 pKa = 6.52DD268 pKa = 4.06ILTMADD274 pKa = 2.98TDD276 pKa = 3.75LTEE279 pKa = 4.43RR280 pKa = 11.84FAAGKK285 pKa = 10.18GAFSLGNDD293 pKa = 3.57PSVMLTCLSLNPDD306 pKa = 2.75ARR308 pKa = 11.84FAFMPSFASKK318 pKa = 10.57EE319 pKa = 4.1GGKK322 pKa = 8.72EE323 pKa = 3.55HH324 pKa = 7.13VGIGEE329 pKa = 4.51GDD331 pKa = 3.41TFGIWKK337 pKa = 8.62DD338 pKa = 3.41TGNADD343 pKa = 3.39AAKK346 pKa = 10.67LFLEE350 pKa = 4.48YY351 pKa = 9.53MARR354 pKa = 11.84PEE356 pKa = 4.13VAQEE360 pKa = 3.69MNAATGKK367 pKa = 10.06ISCLKK372 pKa = 9.54STMEE376 pKa = 4.23IDD378 pKa = 3.74DD379 pKa = 4.27SYY381 pKa = 11.6GLQVFQEE388 pKa = 4.34MKK390 pKa = 10.12EE391 pKa = 3.93KK392 pKa = 10.92CADD395 pKa = 3.41CDD397 pKa = 3.29IFYY400 pKa = 10.88EE401 pKa = 4.24NLWDD405 pKa = 3.86RR406 pKa = 11.84QYY408 pKa = 10.45MPSGMWSIFGNASDD422 pKa = 4.64MLFDD426 pKa = 5.85DD427 pKa = 5.48DD428 pKa = 5.84SEE430 pKa = 5.42AGQDD434 pKa = 3.57EE435 pKa = 4.87VIEE438 pKa = 4.12YY439 pKa = 10.85LLEE442 pKa = 4.39NYY444 pKa = 9.8QDD446 pKa = 4.74LYY448 pKa = 11.56DD449 pKa = 4.3AAQEE453 pKa = 4.29GG454 pKa = 3.97
MM1 pKa = 7.5SMKK4 pKa = 10.55KK5 pKa = 10.16LGSILLSTAMVMSLAACGSQNTSAPAATTDD35 pKa = 3.54DD36 pKa = 3.86SATKK40 pKa = 10.27EE41 pKa = 4.07EE42 pKa = 4.23AVAPAEE48 pKa = 4.28SAVAADD54 pKa = 3.94TDD56 pKa = 4.11VSGSTLEE63 pKa = 4.09VAVTYY68 pKa = 10.21TGDD71 pKa = 3.31QATTFQGLVDD81 pKa = 4.02KK82 pKa = 11.21FEE84 pKa = 4.21EE85 pKa = 4.88EE86 pKa = 4.25YY87 pKa = 11.23GVTVNIAEE95 pKa = 4.29YY96 pKa = 11.04GDD98 pKa = 5.13DD99 pKa = 4.15YY100 pKa = 11.87EE101 pKa = 4.53NTLKK105 pKa = 9.92TRR107 pKa = 11.84MASNEE112 pKa = 4.09LPDD115 pKa = 3.65VFQTHH120 pKa = 5.39GWSLLRR126 pKa = 11.84YY127 pKa = 9.47KK128 pKa = 10.27EE129 pKa = 4.11YY130 pKa = 11.16LMNLQDD136 pKa = 4.08QPWVSDD142 pKa = 3.66YY143 pKa = 11.59DD144 pKa = 3.88EE145 pKa = 4.79SALGVIQDD153 pKa = 4.5DD154 pKa = 4.02DD155 pKa = 4.0GAIYY159 pKa = 11.11VLMISEE165 pKa = 5.79LINGTLVNLDD175 pKa = 3.44ACDD178 pKa = 3.42AAGVDD183 pKa = 4.65PYY185 pKa = 11.29AIHH188 pKa = 6.29TWDD191 pKa = 5.19DD192 pKa = 3.41FTEE195 pKa = 4.38ACAKK199 pKa = 10.17IKK201 pKa = 10.12TSGLTPLNVVSNPGLLANFAGTFVSYY227 pKa = 10.01EE228 pKa = 3.92GEE230 pKa = 4.11MFEE233 pKa = 6.1DD234 pKa = 3.67SAAMLDD240 pKa = 5.56GSFDD244 pKa = 3.31WQDD247 pKa = 3.44YY248 pKa = 10.38KK249 pKa = 11.61DD250 pKa = 3.91SLVKK254 pKa = 10.65YY255 pKa = 6.7MANWIDD261 pKa = 2.94SGYY264 pKa = 10.79YY265 pKa = 9.74YY266 pKa = 10.97DD267 pKa = 6.52DD268 pKa = 4.06ILTMADD274 pKa = 2.98TDD276 pKa = 3.75LTEE279 pKa = 4.43RR280 pKa = 11.84FAAGKK285 pKa = 10.18GAFSLGNDD293 pKa = 3.57PSVMLTCLSLNPDD306 pKa = 2.75ARR308 pKa = 11.84FAFMPSFASKK318 pKa = 10.57EE319 pKa = 4.1GGKK322 pKa = 8.72EE323 pKa = 3.55HH324 pKa = 7.13VGIGEE329 pKa = 4.51GDD331 pKa = 3.41TFGIWKK337 pKa = 8.62DD338 pKa = 3.41TGNADD343 pKa = 3.39AAKK346 pKa = 10.67LFLEE350 pKa = 4.48YY351 pKa = 9.53MARR354 pKa = 11.84PEE356 pKa = 4.13VAQEE360 pKa = 3.69MNAATGKK367 pKa = 10.06ISCLKK372 pKa = 9.54STMEE376 pKa = 4.23IDD378 pKa = 3.74DD379 pKa = 4.27SYY381 pKa = 11.6GLQVFQEE388 pKa = 4.34MKK390 pKa = 10.12EE391 pKa = 3.93KK392 pKa = 10.92CADD395 pKa = 3.41CDD397 pKa = 3.29IFYY400 pKa = 10.88EE401 pKa = 4.24NLWDD405 pKa = 3.86RR406 pKa = 11.84QYY408 pKa = 10.45MPSGMWSIFGNASDD422 pKa = 4.64MLFDD426 pKa = 5.85DD427 pKa = 5.48DD428 pKa = 5.84SEE430 pKa = 5.42AGQDD434 pKa = 3.57EE435 pKa = 4.87VIEE438 pKa = 4.12YY439 pKa = 10.85LLEE442 pKa = 4.39NYY444 pKa = 9.8QDD446 pKa = 4.74LYY448 pKa = 11.56DD449 pKa = 4.3AAQEE453 pKa = 4.29GG454 pKa = 3.97
Molecular weight: 49.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5H0Z3|R5H0Z3_9FIRM Aldose 1-epimerase OS=Firmicutes bacterium CAG:24 OX=1263012 GN=BN555_01445 PE=3 SV=1
MM1 pKa = 7.69TIQKK5 pKa = 9.78RR6 pKa = 11.84FFIIFFRR13 pKa = 11.84IFFYY17 pKa = 10.94VSPSSDD23 pKa = 2.61RR24 pKa = 11.84RR25 pKa = 11.84QRR27 pKa = 11.84VHH29 pKa = 6.09NCNIIYY35 pKa = 10.05RR36 pKa = 11.84LCFSKK41 pKa = 11.07NSFEE45 pKa = 4.14RR46 pKa = 11.84MSVLFRR52 pKa = 11.84QRR54 pKa = 11.84IKK56 pKa = 10.81RR57 pKa = 11.84IDD59 pKa = 3.34FEE61 pKa = 4.45YY62 pKa = 9.91FIRR65 pKa = 11.84MLQKK69 pKa = 10.12FQTQIQCQIAFSASSRR85 pKa = 11.84RR86 pKa = 11.84LYY88 pKa = 10.58KK89 pKa = 10.11INTVFFCSLQIFFCFQKK106 pKa = 10.72EE107 pKa = 4.29LISEE111 pKa = 4.85DD112 pKa = 3.21IDD114 pKa = 4.06LLFTIPFLSRR124 pKa = 11.84ITFFSILPNRR134 pKa = 11.84FHH136 pKa = 7.12QRR138 pKa = 11.84RR139 pKa = 11.84TLINLPEE146 pKa = 4.6FGSVSLSKK154 pKa = 9.74TIHH157 pKa = 6.24LKK159 pKa = 9.15STVKK163 pKa = 8.85FCRR166 pKa = 11.84RR167 pKa = 11.84IVVTGYY173 pKa = 8.51QNRR176 pKa = 11.84IFQTDD181 pKa = 2.89KK182 pKa = 10.82KK183 pKa = 9.89IVYY186 pKa = 9.98NIVFCSPKK194 pKa = 9.92RR195 pKa = 11.84FSAHH199 pKa = 5.97KK200 pKa = 9.63VLIYY204 pKa = 8.08TFRR207 pKa = 11.84YY208 pKa = 8.9RR209 pKa = 11.84DD210 pKa = 3.74LVKK213 pKa = 10.39KK214 pKa = 10.36AQLGKK219 pKa = 9.95SRR221 pKa = 11.84NRR223 pKa = 11.84FFFPLSQQFLCFQLKK238 pKa = 9.13VFQICFHH245 pKa = 6.49RR246 pKa = 11.84FEE248 pKa = 4.45IAVMFCDD255 pKa = 5.05YY256 pKa = 10.65IRR258 pKa = 11.84QFPSKK263 pKa = 10.17CNQRR267 pKa = 11.84FLLLFFHH274 pKa = 7.55FIEE277 pKa = 5.06TIFNFII283 pKa = 4.16
MM1 pKa = 7.69TIQKK5 pKa = 9.78RR6 pKa = 11.84FFIIFFRR13 pKa = 11.84IFFYY17 pKa = 10.94VSPSSDD23 pKa = 2.61RR24 pKa = 11.84RR25 pKa = 11.84QRR27 pKa = 11.84VHH29 pKa = 6.09NCNIIYY35 pKa = 10.05RR36 pKa = 11.84LCFSKK41 pKa = 11.07NSFEE45 pKa = 4.14RR46 pKa = 11.84MSVLFRR52 pKa = 11.84QRR54 pKa = 11.84IKK56 pKa = 10.81RR57 pKa = 11.84IDD59 pKa = 3.34FEE61 pKa = 4.45YY62 pKa = 9.91FIRR65 pKa = 11.84MLQKK69 pKa = 10.12FQTQIQCQIAFSASSRR85 pKa = 11.84RR86 pKa = 11.84LYY88 pKa = 10.58KK89 pKa = 10.11INTVFFCSLQIFFCFQKK106 pKa = 10.72EE107 pKa = 4.29LISEE111 pKa = 4.85DD112 pKa = 3.21IDD114 pKa = 4.06LLFTIPFLSRR124 pKa = 11.84ITFFSILPNRR134 pKa = 11.84FHH136 pKa = 7.12QRR138 pKa = 11.84RR139 pKa = 11.84TLINLPEE146 pKa = 4.6FGSVSLSKK154 pKa = 9.74TIHH157 pKa = 6.24LKK159 pKa = 9.15STVKK163 pKa = 8.85FCRR166 pKa = 11.84RR167 pKa = 11.84IVVTGYY173 pKa = 8.51QNRR176 pKa = 11.84IFQTDD181 pKa = 2.89KK182 pKa = 10.82KK183 pKa = 9.89IVYY186 pKa = 9.98NIVFCSPKK194 pKa = 9.92RR195 pKa = 11.84FSAHH199 pKa = 5.97KK200 pKa = 9.63VLIYY204 pKa = 8.08TFRR207 pKa = 11.84YY208 pKa = 8.9RR209 pKa = 11.84DD210 pKa = 3.74LVKK213 pKa = 10.39KK214 pKa = 10.36AQLGKK219 pKa = 9.95SRR221 pKa = 11.84NRR223 pKa = 11.84FFFPLSQQFLCFQLKK238 pKa = 9.13VFQICFHH245 pKa = 6.49RR246 pKa = 11.84FEE248 pKa = 4.45IAVMFCDD255 pKa = 5.05YY256 pKa = 10.65IRR258 pKa = 11.84QFPSKK263 pKa = 10.17CNQRR267 pKa = 11.84FLLLFFHH274 pKa = 7.55FIEE277 pKa = 5.06TIFNFII283 pKa = 4.16
Molecular weight: 34.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
809563 |
29 |
1630 |
321.5 |
35.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.225 ± 0.053 | 1.629 ± 0.018 |
5.485 ± 0.04 | 7.59 ± 0.055 |
4.18 ± 0.032 | 7.282 ± 0.039 |
1.788 ± 0.021 | 6.565 ± 0.042 |
6.235 ± 0.031 | 9.463 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.985 ± 0.022 | 3.832 ± 0.03 |
3.451 ± 0.026 | 3.65 ± 0.028 |
4.785 ± 0.041 | 5.628 ± 0.036 |
5.266 ± 0.028 | 6.904 ± 0.041 |
1.088 ± 0.02 | 3.967 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
