
Streptococcus phage CHPC1083
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Brussowvirus; unclassified Brussowvirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
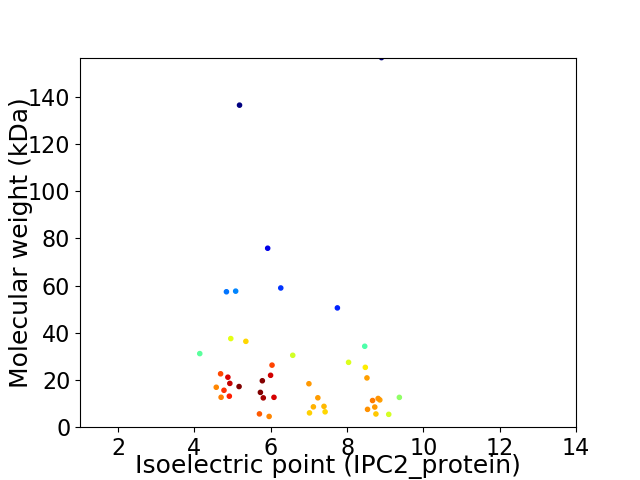
Virtual 2D-PAGE plot for 45 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
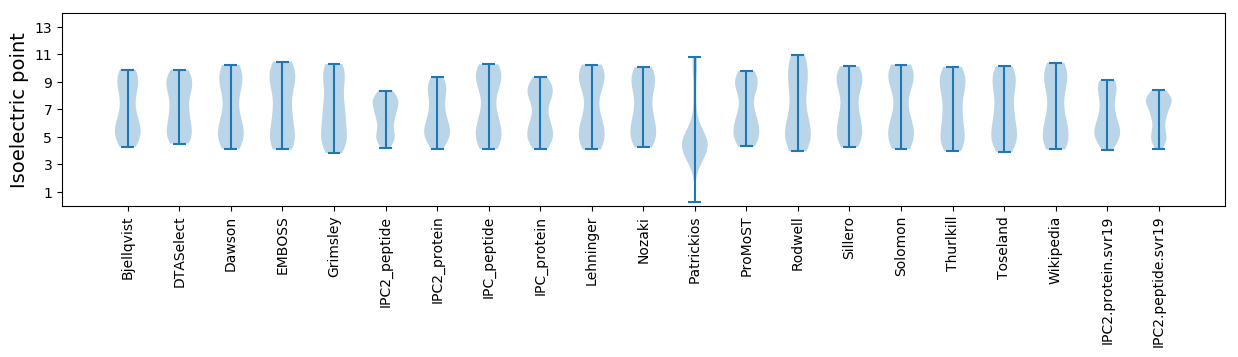
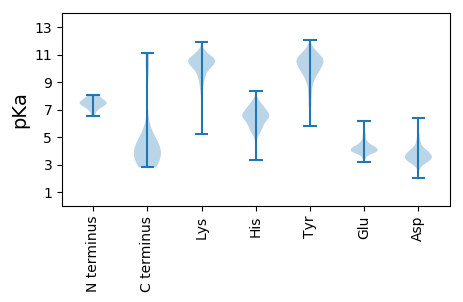
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G8FAT7|A0A3G8FAT7_9CAUD Uncharacterized protein OS=Streptococcus phage CHPC1083 OX=2365024 GN=CHPC1083_0019 PE=4 SV=1
MM1 pKa = 7.56SVPQSIVNWFVIHH14 pKa = 7.14RR15 pKa = 11.84NLLTYY20 pKa = 11.39SMFGSRR26 pKa = 11.84NGSDD30 pKa = 3.03GTADD34 pKa = 3.56CSGSMSQALKK44 pKa = 10.76DD45 pKa = 3.46AGIPIQGLPSTVTLGQQLAKK65 pKa = 10.58NGFYY69 pKa = 10.18RR70 pKa = 11.84VSINQDD76 pKa = 2.24WDD78 pKa = 3.21ASTGDD83 pKa = 3.44IVMMSWGADD92 pKa = 3.23MSQSGGAGGHH102 pKa = 5.81VGVMMDD108 pKa = 3.26SVNFISCDD116 pKa = 3.3YY117 pKa = 8.82STQGAVGQAINTYY130 pKa = 8.91PWNDD134 pKa = 3.08YY135 pKa = 10.16YY136 pKa = 11.52AANKK140 pKa = 9.07PNYY143 pKa = 9.09IEE145 pKa = 3.75VWRR148 pKa = 11.84YY149 pKa = 10.19AEE151 pKa = 5.22SAPQTNNQANTAVVPQQKK169 pKa = 10.42AYY171 pKa = 11.13YY172 pKa = 8.27EE173 pKa = 4.42ANDD176 pKa = 3.44VQFVNGIWQIKK187 pKa = 9.47CDD189 pKa = 4.14YY190 pKa = 9.88LCPIGFNYY198 pKa = 8.74FQNGVPVTMVNWVDD212 pKa = 3.69KK213 pKa = 11.03DD214 pKa = 4.24GNDD217 pKa = 3.97LPDD220 pKa = 4.26GADD223 pKa = 3.29QEE225 pKa = 5.13FKK227 pKa = 11.28AGMFFSFAGDD237 pKa = 3.56EE238 pKa = 4.15NNITDD243 pKa = 3.82TGEE246 pKa = 3.65GGYY249 pKa = 10.61YY250 pKa = 9.76GGYY253 pKa = 8.0YY254 pKa = 9.41YY255 pKa = 10.76RR256 pKa = 11.84RR257 pKa = 11.84FEE259 pKa = 4.06FGQFGTVWLSCWNKK273 pKa = 10.66DD274 pKa = 3.89DD275 pKa = 5.14LVNYY279 pKa = 7.38YY280 pKa = 8.26QQ281 pKa = 4.22
MM1 pKa = 7.56SVPQSIVNWFVIHH14 pKa = 7.14RR15 pKa = 11.84NLLTYY20 pKa = 11.39SMFGSRR26 pKa = 11.84NGSDD30 pKa = 3.03GTADD34 pKa = 3.56CSGSMSQALKK44 pKa = 10.76DD45 pKa = 3.46AGIPIQGLPSTVTLGQQLAKK65 pKa = 10.58NGFYY69 pKa = 10.18RR70 pKa = 11.84VSINQDD76 pKa = 2.24WDD78 pKa = 3.21ASTGDD83 pKa = 3.44IVMMSWGADD92 pKa = 3.23MSQSGGAGGHH102 pKa = 5.81VGVMMDD108 pKa = 3.26SVNFISCDD116 pKa = 3.3YY117 pKa = 8.82STQGAVGQAINTYY130 pKa = 8.91PWNDD134 pKa = 3.08YY135 pKa = 10.16YY136 pKa = 11.52AANKK140 pKa = 9.07PNYY143 pKa = 9.09IEE145 pKa = 3.75VWRR148 pKa = 11.84YY149 pKa = 10.19AEE151 pKa = 5.22SAPQTNNQANTAVVPQQKK169 pKa = 10.42AYY171 pKa = 11.13YY172 pKa = 8.27EE173 pKa = 4.42ANDD176 pKa = 3.44VQFVNGIWQIKK187 pKa = 9.47CDD189 pKa = 4.14YY190 pKa = 9.88LCPIGFNYY198 pKa = 8.74FQNGVPVTMVNWVDD212 pKa = 3.69KK213 pKa = 11.03DD214 pKa = 4.24GNDD217 pKa = 3.97LPDD220 pKa = 4.26GADD223 pKa = 3.29QEE225 pKa = 5.13FKK227 pKa = 11.28AGMFFSFAGDD237 pKa = 3.56EE238 pKa = 4.15NNITDD243 pKa = 3.82TGEE246 pKa = 3.65GGYY249 pKa = 10.61YY250 pKa = 9.76GGYY253 pKa = 8.0YY254 pKa = 9.41YY255 pKa = 10.76RR256 pKa = 11.84RR257 pKa = 11.84FEE259 pKa = 4.06FGQFGTVWLSCWNKK273 pKa = 10.66DD274 pKa = 3.89DD275 pKa = 5.14LVNYY279 pKa = 7.38YY280 pKa = 8.26QQ281 pKa = 4.22
Molecular weight: 31.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G8FAP5|A0A3G8FAP5_9CAUD Uncharacterized protein OS=Streptococcus phage CHPC1083 OX=2365024 GN=CHPC1083_0032 PE=4 SV=1
MM1 pKa = 7.83ADD3 pKa = 3.75FGVSLLEE10 pKa = 4.01ARR12 pKa = 11.84RR13 pKa = 11.84MTLKK17 pKa = 10.49EE18 pKa = 3.53MKK20 pKa = 9.92LYY22 pKa = 10.32QKK24 pKa = 10.55AYY26 pKa = 9.84KK27 pKa = 10.03KK28 pKa = 10.52RR29 pKa = 11.84FLNKK33 pKa = 9.43EE34 pKa = 3.69RR35 pKa = 11.84EE36 pKa = 4.36IYY38 pKa = 9.68QLAYY42 pKa = 10.87LNRR45 pKa = 11.84LANATTKK52 pKa = 10.62DD53 pKa = 3.03GKK55 pKa = 10.64KK56 pKa = 10.46YY57 pKa = 10.49YY58 pKa = 10.19FEE60 pKa = 5.48KK61 pKa = 10.83FDD63 pKa = 3.95DD64 pKa = 4.49FYY66 pKa = 11.56NAKK69 pKa = 9.54EE70 pKa = 3.83RR71 pKa = 11.84ARR73 pKa = 11.84EE74 pKa = 3.84VLGEE78 pKa = 4.35KK79 pKa = 8.61ITNSKK84 pKa = 10.18LLEE87 pKa = 3.98RR88 pKa = 11.84ARR90 pKa = 11.84NNLNYY95 pKa = 10.06KK96 pKa = 9.85KK97 pKa = 10.46EE98 pKa = 3.94RR99 pKa = 11.84GLLDD103 pKa = 3.22GRR105 pKa = 4.46
MM1 pKa = 7.83ADD3 pKa = 3.75FGVSLLEE10 pKa = 4.01ARR12 pKa = 11.84RR13 pKa = 11.84MTLKK17 pKa = 10.49EE18 pKa = 3.53MKK20 pKa = 9.92LYY22 pKa = 10.32QKK24 pKa = 10.55AYY26 pKa = 9.84KK27 pKa = 10.03KK28 pKa = 10.52RR29 pKa = 11.84FLNKK33 pKa = 9.43EE34 pKa = 3.69RR35 pKa = 11.84EE36 pKa = 4.36IYY38 pKa = 9.68QLAYY42 pKa = 10.87LNRR45 pKa = 11.84LANATTKK52 pKa = 10.62DD53 pKa = 3.03GKK55 pKa = 10.64KK56 pKa = 10.46YY57 pKa = 10.49YY58 pKa = 10.19FEE60 pKa = 5.48KK61 pKa = 10.83FDD63 pKa = 3.95DD64 pKa = 4.49FYY66 pKa = 11.56NAKK69 pKa = 9.54EE70 pKa = 3.83RR71 pKa = 11.84ARR73 pKa = 11.84EE74 pKa = 3.84VLGEE78 pKa = 4.35KK79 pKa = 8.61ITNSKK84 pKa = 10.18LLEE87 pKa = 3.98RR88 pKa = 11.84ARR90 pKa = 11.84NNLNYY95 pKa = 10.06KK96 pKa = 9.85KK97 pKa = 10.46EE98 pKa = 3.94RR99 pKa = 11.84GLLDD103 pKa = 3.22GRR105 pKa = 4.46
Molecular weight: 12.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10987 |
41 |
1545 |
244.2 |
27.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.709 ± 1.191 | 0.519 ± 0.108 |
6.189 ± 0.515 | 6.681 ± 0.834 |
4.187 ± 0.166 | 6.799 ± 0.545 |
1.329 ± 0.197 | 6.863 ± 0.401 |
7.955 ± 0.671 | 7.664 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.321 ± 0.296 | 5.925 ± 0.466 |
2.849 ± 0.275 | 3.95 ± 0.248 |
4.169 ± 0.425 | 6.872 ± 0.677 |
6.435 ± 0.384 | 6.635 ± 0.272 |
1.156 ± 0.188 | 3.795 ± 0.496 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
