
Klebsiella phage vB_Kpn_Chronis
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
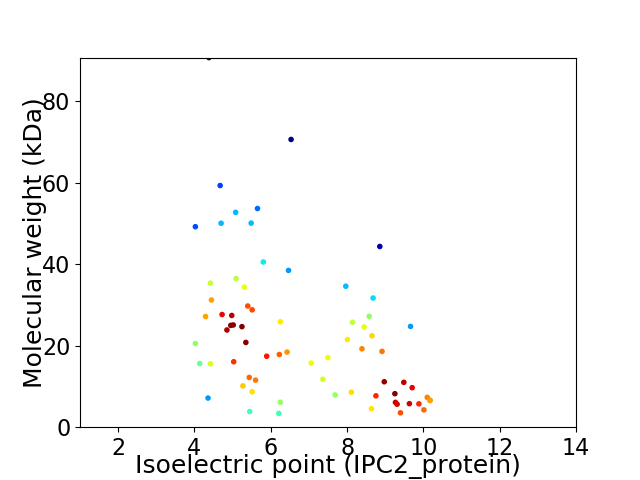
Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B9MVE9|A0A5B9MVE9_9CAUD Putative conjugal transfer protein OS=Klebsiella phage vB_Kpn_Chronis OX=2591378 GN=CHRON_30 PE=4 SV=1
MM1 pKa = 7.7SDD3 pKa = 4.0LPVSYY8 pKa = 9.92TSAGPVPLTAEE19 pKa = 4.16EE20 pKa = 4.41LRR22 pKa = 11.84AQLVSQAIALSPGLTTDD39 pKa = 4.11LPGSLIEE46 pKa = 5.3DD47 pKa = 3.6VASTDD52 pKa = 3.14VGALIVCDD60 pKa = 3.6QARR63 pKa = 11.84VDD65 pKa = 5.4LINSVGPLKK74 pKa = 11.07ANLAMLEE81 pKa = 4.38LLAQQAGIPGQKK93 pKa = 8.44TAGTTTVPVQFSGPAGFVIPQGFIVSDD120 pKa = 3.52GTYY123 pKa = 9.19TYY125 pKa = 10.59SVSDD129 pKa = 3.34ATIISSSGVSASVSCEE145 pKa = 3.61GTEE148 pKa = 3.97TGTWAVPVNTVNQIISSLPSDD169 pKa = 3.66VTITCTNPIAGTPGADD185 pKa = 3.6PEE187 pKa = 4.52TNYY190 pKa = 10.29QFRR193 pKa = 11.84DD194 pKa = 3.97RR195 pKa = 11.84VWQAQMATVQGYY207 pKa = 8.99PGFIRR212 pKa = 11.84QYY214 pKa = 9.62LTSLDD219 pKa = 3.63NVQARR224 pKa = 11.84LVSVIQDD231 pKa = 3.21GDD233 pKa = 2.97KK234 pKa = 10.34WIVMCAGGDD243 pKa = 3.21IYY245 pKa = 11.02DD246 pKa = 3.88IAGALYY252 pKa = 10.24KK253 pKa = 10.58SAGDD257 pKa = 3.3ISRR260 pKa = 11.84LKK262 pKa = 10.58GCSLNVTGITNANPGVVSTDD282 pKa = 3.2LTHH285 pKa = 7.35GYY287 pKa = 8.81TDD289 pKa = 3.62GQVIRR294 pKa = 11.84ITGVTGMTGINDD306 pKa = 3.45VPLTVTVLSPHH317 pKa = 6.03TFSIGIDD324 pKa = 3.62TTSSGTWGGGGEE336 pKa = 4.11VTPNVRR342 pKa = 11.84NNTVTVNDD350 pKa = 3.6WPDD353 pKa = 2.96NYY355 pKa = 10.33VIPFVTPLLQRR366 pKa = 11.84VTVTYY371 pKa = 10.02QWGTEE376 pKa = 4.12SVNYY380 pKa = 8.89LTDD383 pKa = 3.2ATIASLVSAPTIQYY397 pKa = 10.5VNGIFAGKK405 pKa = 8.53PLNVNNLKK413 pKa = 10.63DD414 pKa = 3.74AFLQAINSTIDD425 pKa = 2.98MGLISTLNVVVTVNGVITPPDD446 pKa = 3.79AGTNIISGDD455 pKa = 3.5KK456 pKa = 10.4FSYY459 pKa = 10.1FYY461 pKa = 10.44IASDD465 pKa = 3.86GVIVTGAA472 pKa = 3.16
MM1 pKa = 7.7SDD3 pKa = 4.0LPVSYY8 pKa = 9.92TSAGPVPLTAEE19 pKa = 4.16EE20 pKa = 4.41LRR22 pKa = 11.84AQLVSQAIALSPGLTTDD39 pKa = 4.11LPGSLIEE46 pKa = 5.3DD47 pKa = 3.6VASTDD52 pKa = 3.14VGALIVCDD60 pKa = 3.6QARR63 pKa = 11.84VDD65 pKa = 5.4LINSVGPLKK74 pKa = 11.07ANLAMLEE81 pKa = 4.38LLAQQAGIPGQKK93 pKa = 8.44TAGTTTVPVQFSGPAGFVIPQGFIVSDD120 pKa = 3.52GTYY123 pKa = 9.19TYY125 pKa = 10.59SVSDD129 pKa = 3.34ATIISSSGVSASVSCEE145 pKa = 3.61GTEE148 pKa = 3.97TGTWAVPVNTVNQIISSLPSDD169 pKa = 3.66VTITCTNPIAGTPGADD185 pKa = 3.6PEE187 pKa = 4.52TNYY190 pKa = 10.29QFRR193 pKa = 11.84DD194 pKa = 3.97RR195 pKa = 11.84VWQAQMATVQGYY207 pKa = 8.99PGFIRR212 pKa = 11.84QYY214 pKa = 9.62LTSLDD219 pKa = 3.63NVQARR224 pKa = 11.84LVSVIQDD231 pKa = 3.21GDD233 pKa = 2.97KK234 pKa = 10.34WIVMCAGGDD243 pKa = 3.21IYY245 pKa = 11.02DD246 pKa = 3.88IAGALYY252 pKa = 10.24KK253 pKa = 10.58SAGDD257 pKa = 3.3ISRR260 pKa = 11.84LKK262 pKa = 10.58GCSLNVTGITNANPGVVSTDD282 pKa = 3.2LTHH285 pKa = 7.35GYY287 pKa = 8.81TDD289 pKa = 3.62GQVIRR294 pKa = 11.84ITGVTGMTGINDD306 pKa = 3.45VPLTVTVLSPHH317 pKa = 6.03TFSIGIDD324 pKa = 3.62TTSSGTWGGGGEE336 pKa = 4.11VTPNVRR342 pKa = 11.84NNTVTVNDD350 pKa = 3.6WPDD353 pKa = 2.96NYY355 pKa = 10.33VIPFVTPLLQRR366 pKa = 11.84VTVTYY371 pKa = 10.02QWGTEE376 pKa = 4.12SVNYY380 pKa = 8.89LTDD383 pKa = 3.2ATIASLVSAPTIQYY397 pKa = 10.5VNGIFAGKK405 pKa = 8.53PLNVNNLKK413 pKa = 10.63DD414 pKa = 3.74AFLQAINSTIDD425 pKa = 2.98MGLISTLNVVVTVNGVITPPDD446 pKa = 3.79AGTNIISGDD455 pKa = 3.5KK456 pKa = 10.4FSYY459 pKa = 10.1FYY461 pKa = 10.44IASDD465 pKa = 3.86GVIVTGAA472 pKa = 3.16
Molecular weight: 49.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9MSR3|A0A5B9MSR3_9CAUD Putative YlcG family protein OS=Klebsiella phage vB_Kpn_Chronis OX=2591378 GN=CHRON_64 PE=4 SV=1
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 8.26PARR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.32CAHH11 pKa = 5.64CRR13 pKa = 11.84EE14 pKa = 4.41WFHH17 pKa = 6.84PARR20 pKa = 11.84EE21 pKa = 4.26GQVVCSFEE29 pKa = 4.09CASAIGKK36 pKa = 8.72KK37 pKa = 7.2QTAKK41 pKa = 10.56ARR43 pKa = 11.84EE44 pKa = 3.79AAKK47 pKa = 10.37ARR49 pKa = 11.84EE50 pKa = 4.36VKK52 pKa = 10.06RR53 pKa = 11.84QRR55 pKa = 11.84EE56 pKa = 4.22SEE58 pKa = 4.0KK59 pKa = 10.31EE60 pKa = 3.66GRR62 pKa = 11.84QRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84AKK68 pKa = 10.5RR69 pKa = 11.84EE70 pKa = 3.81SFKK73 pKa = 10.93TKK75 pKa = 10.43AQWDD79 pKa = 4.06KK80 pKa = 10.73EE81 pKa = 4.19AQSAFNRR88 pKa = 11.84YY89 pKa = 8.22IRR91 pKa = 11.84IRR93 pKa = 11.84DD94 pKa = 3.38EE95 pKa = 4.24GKK97 pKa = 9.03PCVSCGSPLIGKK109 pKa = 9.58RR110 pKa = 11.84NYY112 pKa = 9.23LTGSAIDD119 pKa = 3.57ASHH122 pKa = 5.58YY123 pKa = 9.46RR124 pKa = 11.84SRR126 pKa = 11.84GAASHH131 pKa = 6.67LKK133 pKa = 9.12FNVFNVHH140 pKa = 6.17SACTRR145 pKa = 11.84CNRR148 pKa = 11.84QLSGNAVEE156 pKa = 4.08YY157 pKa = 10.25RR158 pKa = 11.84IRR160 pKa = 11.84LIEE163 pKa = 4.66RR164 pKa = 11.84IGLDD168 pKa = 2.76RR169 pKa = 11.84VEE171 pKa = 5.22RR172 pKa = 11.84LEE174 pKa = 3.99TDD176 pKa = 3.21NEE178 pKa = 3.83PRR180 pKa = 11.84RR181 pKa = 11.84FDD183 pKa = 3.05IPYY186 pKa = 8.71LQRR189 pKa = 11.84IKK191 pKa = 11.0SIFTRR196 pKa = 11.84RR197 pKa = 11.84ARR199 pKa = 11.84ALEE202 pKa = 3.86KK203 pKa = 10.4RR204 pKa = 11.84RR205 pKa = 11.84ARR207 pKa = 11.84HH208 pKa = 5.15QEE210 pKa = 3.52AAA212 pKa = 3.66
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 8.26PARR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.32CAHH11 pKa = 5.64CRR13 pKa = 11.84EE14 pKa = 4.41WFHH17 pKa = 6.84PARR20 pKa = 11.84EE21 pKa = 4.26GQVVCSFEE29 pKa = 4.09CASAIGKK36 pKa = 8.72KK37 pKa = 7.2QTAKK41 pKa = 10.56ARR43 pKa = 11.84EE44 pKa = 3.79AAKK47 pKa = 10.37ARR49 pKa = 11.84EE50 pKa = 4.36VKK52 pKa = 10.06RR53 pKa = 11.84QRR55 pKa = 11.84EE56 pKa = 4.22SEE58 pKa = 4.0KK59 pKa = 10.31EE60 pKa = 3.66GRR62 pKa = 11.84QRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84AKK68 pKa = 10.5RR69 pKa = 11.84EE70 pKa = 3.81SFKK73 pKa = 10.93TKK75 pKa = 10.43AQWDD79 pKa = 4.06KK80 pKa = 10.73EE81 pKa = 4.19AQSAFNRR88 pKa = 11.84YY89 pKa = 8.22IRR91 pKa = 11.84IRR93 pKa = 11.84DD94 pKa = 3.38EE95 pKa = 4.24GKK97 pKa = 9.03PCVSCGSPLIGKK109 pKa = 9.58RR110 pKa = 11.84NYY112 pKa = 9.23LTGSAIDD119 pKa = 3.57ASHH122 pKa = 5.58YY123 pKa = 9.46RR124 pKa = 11.84SRR126 pKa = 11.84GAASHH131 pKa = 6.67LKK133 pKa = 9.12FNVFNVHH140 pKa = 6.17SACTRR145 pKa = 11.84CNRR148 pKa = 11.84QLSGNAVEE156 pKa = 4.08YY157 pKa = 10.25RR158 pKa = 11.84IRR160 pKa = 11.84LIEE163 pKa = 4.66RR164 pKa = 11.84IGLDD168 pKa = 2.76RR169 pKa = 11.84VEE171 pKa = 5.22RR172 pKa = 11.84LEE174 pKa = 3.99TDD176 pKa = 3.21NEE178 pKa = 3.83PRR180 pKa = 11.84RR181 pKa = 11.84FDD183 pKa = 3.05IPYY186 pKa = 8.71LQRR189 pKa = 11.84IKK191 pKa = 11.0SIFTRR196 pKa = 11.84RR197 pKa = 11.84ARR199 pKa = 11.84ALEE202 pKa = 3.86KK203 pKa = 10.4RR204 pKa = 11.84RR205 pKa = 11.84ARR207 pKa = 11.84HH208 pKa = 5.15QEE210 pKa = 3.52AAA212 pKa = 3.66
Molecular weight: 24.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14677 |
31 |
849 |
206.7 |
22.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.13 ± 0.353 | 1.138 ± 0.121 |
6.03 ± 0.27 | 5.812 ± 0.405 |
3.509 ± 0.182 | 7.781 ± 0.4 |
1.492 ± 0.126 | 5.969 ± 0.221 |
5.246 ± 0.358 | 7.856 ± 0.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.909 ± 0.134 | 4.756 ± 0.234 |
4.19 ± 0.196 | 4.183 ± 0.193 |
5.723 ± 0.346 | 6.82 ± 0.263 |
6.323 ± 0.403 | 6.343 ± 0.254 |
1.574 ± 0.118 | 3.216 ± 0.165 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
