
Lake Sarah-associated circular virus-22
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
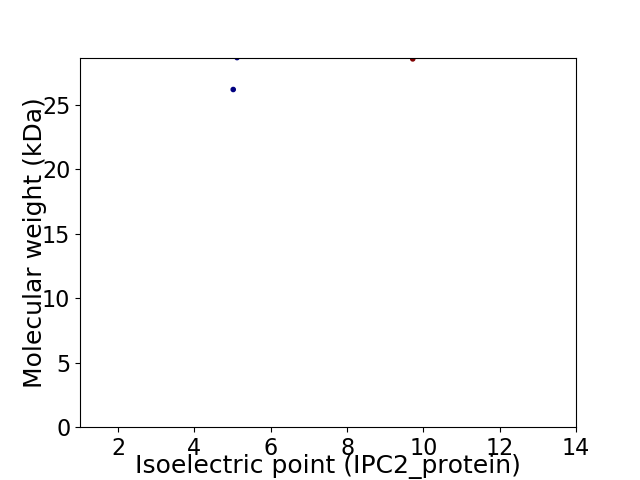
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
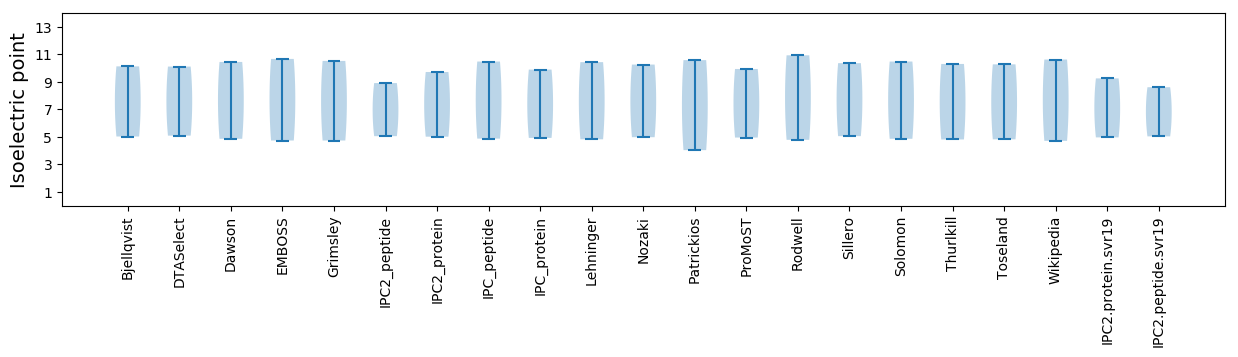
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A140AQM3|A0A140AQM3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-22 OX=1685749 PE=4 SV=1
MM1 pKa = 7.62SEE3 pKa = 5.24RR4 pKa = 11.84ATCWSVTINNPIPADD19 pKa = 3.79EE20 pKa = 5.26DD21 pKa = 3.76NIAQANQRR29 pKa = 11.84SGWKK33 pKa = 10.09VEE35 pKa = 4.07GQLEE39 pKa = 4.12QGEE42 pKa = 4.87NGTQHH47 pKa = 5.69YY48 pKa = 10.38QLMVKK53 pKa = 9.09TPQIRR58 pKa = 11.84FSQIKK63 pKa = 9.0KK64 pKa = 8.62QFPRR68 pKa = 11.84AHH70 pKa = 6.34IEE72 pKa = 3.6VARR75 pKa = 11.84NVKK78 pKa = 9.55ALKK81 pKa = 10.33EE82 pKa = 4.0YY83 pKa = 9.39VHH85 pKa = 7.44KK86 pKa = 10.72DD87 pKa = 3.04DD88 pKa = 4.39TKK90 pKa = 10.61IGEE93 pKa = 4.06IPVNDD98 pKa = 4.69KK99 pKa = 10.95YY100 pKa = 10.93PTMTAFWEE108 pKa = 4.67LIYY111 pKa = 10.45DD112 pKa = 3.31IHH114 pKa = 6.3EE115 pKa = 4.02QLKK118 pKa = 10.55RR119 pKa = 11.84DD120 pKa = 3.38IDD122 pKa = 3.8YY123 pKa = 10.38MGIEE127 pKa = 4.29TPEE130 pKa = 3.88EE131 pKa = 3.88RR132 pKa = 11.84LVLLDD137 pKa = 3.18QCVNLLICQTYY148 pKa = 9.11NVEE151 pKa = 4.33TFGVNPQVRR160 pKa = 11.84SAAKK164 pKa = 9.73KK165 pKa = 8.8YY166 pKa = 10.68LPALFIRR173 pKa = 11.84VEE175 pKa = 3.73KK176 pKa = 10.04MRR178 pKa = 11.84RR179 pKa = 11.84QKK181 pKa = 8.71TVRR184 pKa = 11.84QTEE187 pKa = 4.17LNSAVEE193 pKa = 4.01QNEE196 pKa = 4.2NEE198 pKa = 4.18EE199 pKa = 4.63STSEE203 pKa = 4.12TQSTCEE209 pKa = 4.01TQSTCEE215 pKa = 4.01TDD217 pKa = 3.41EE218 pKa = 5.04SSQSDD223 pKa = 3.4WNDD226 pKa = 2.68
MM1 pKa = 7.62SEE3 pKa = 5.24RR4 pKa = 11.84ATCWSVTINNPIPADD19 pKa = 3.79EE20 pKa = 5.26DD21 pKa = 3.76NIAQANQRR29 pKa = 11.84SGWKK33 pKa = 10.09VEE35 pKa = 4.07GQLEE39 pKa = 4.12QGEE42 pKa = 4.87NGTQHH47 pKa = 5.69YY48 pKa = 10.38QLMVKK53 pKa = 9.09TPQIRR58 pKa = 11.84FSQIKK63 pKa = 9.0KK64 pKa = 8.62QFPRR68 pKa = 11.84AHH70 pKa = 6.34IEE72 pKa = 3.6VARR75 pKa = 11.84NVKK78 pKa = 9.55ALKK81 pKa = 10.33EE82 pKa = 4.0YY83 pKa = 9.39VHH85 pKa = 7.44KK86 pKa = 10.72DD87 pKa = 3.04DD88 pKa = 4.39TKK90 pKa = 10.61IGEE93 pKa = 4.06IPVNDD98 pKa = 4.69KK99 pKa = 10.95YY100 pKa = 10.93PTMTAFWEE108 pKa = 4.67LIYY111 pKa = 10.45DD112 pKa = 3.31IHH114 pKa = 6.3EE115 pKa = 4.02QLKK118 pKa = 10.55RR119 pKa = 11.84DD120 pKa = 3.38IDD122 pKa = 3.8YY123 pKa = 10.38MGIEE127 pKa = 4.29TPEE130 pKa = 3.88EE131 pKa = 3.88RR132 pKa = 11.84LVLLDD137 pKa = 3.18QCVNLLICQTYY148 pKa = 9.11NVEE151 pKa = 4.33TFGVNPQVRR160 pKa = 11.84SAAKK164 pKa = 9.73KK165 pKa = 8.8YY166 pKa = 10.68LPALFIRR173 pKa = 11.84VEE175 pKa = 3.73KK176 pKa = 10.04MRR178 pKa = 11.84RR179 pKa = 11.84QKK181 pKa = 8.71TVRR184 pKa = 11.84QTEE187 pKa = 4.17LNSAVEE193 pKa = 4.01QNEE196 pKa = 4.2NEE198 pKa = 4.18EE199 pKa = 4.63STSEE203 pKa = 4.12TQSTCEE209 pKa = 4.01TQSTCEE215 pKa = 4.01TDD217 pKa = 3.41EE218 pKa = 5.04SSQSDD223 pKa = 3.4WNDD226 pKa = 2.68
Molecular weight: 26.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQM3|A0A140AQM3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-22 OX=1685749 PE=4 SV=1
MM1 pKa = 7.27KK2 pKa = 9.94MKK4 pKa = 10.4RR5 pKa = 11.84APAKK9 pKa = 9.1PRR11 pKa = 11.84APARR15 pKa = 11.84PRR17 pKa = 11.84APARR21 pKa = 11.84RR22 pKa = 11.84TRR24 pKa = 11.84VPRR27 pKa = 11.84AIGMTKK33 pKa = 10.17SGLKK37 pKa = 9.74TYY39 pKa = 9.58TFTFKK44 pKa = 10.42PAPQVFIQAVPTPSSGNTLILSSATGAGTAPLSNGKK80 pKa = 9.21CLTPASSNGLTGFYY94 pKa = 10.76DD95 pKa = 3.67WTCATNFLLADD106 pKa = 4.11LQSAPTYY113 pKa = 10.02TGLFDD118 pKa = 4.03AYY120 pKa = 9.24KK121 pKa = 10.04IKK123 pKa = 10.47KK124 pKa = 7.24ITMNIEE130 pKa = 3.89YY131 pKa = 10.29LNNTSSVNSTGLMPTIYY148 pKa = 10.22AYY150 pKa = 10.61FDD152 pKa = 3.5QDD154 pKa = 3.34DD155 pKa = 4.01SAVPVSVADD164 pKa = 3.18ITRR167 pKa = 11.84RR168 pKa = 11.84QGVKK172 pKa = 10.12KK173 pKa = 10.04FQFGNKK179 pKa = 8.65SKK181 pKa = 9.89TSFSFSLKK189 pKa = 8.54PTVSSALEE197 pKa = 4.31TNSGTLIQAAVQKK210 pKa = 10.64AGWINCTQFAIPHH223 pKa = 5.99YY224 pKa = 8.57GLKK227 pKa = 10.44FAVTDD232 pKa = 4.58LYY234 pKa = 11.39LPGVNAIQQAFRR246 pKa = 11.84INWTYY251 pKa = 9.78VVSYY255 pKa = 10.21RR256 pKa = 11.84SPLRR260 pKa = 11.84NNN262 pKa = 3.27
MM1 pKa = 7.27KK2 pKa = 9.94MKK4 pKa = 10.4RR5 pKa = 11.84APAKK9 pKa = 9.1PRR11 pKa = 11.84APARR15 pKa = 11.84PRR17 pKa = 11.84APARR21 pKa = 11.84RR22 pKa = 11.84TRR24 pKa = 11.84VPRR27 pKa = 11.84AIGMTKK33 pKa = 10.17SGLKK37 pKa = 9.74TYY39 pKa = 9.58TFTFKK44 pKa = 10.42PAPQVFIQAVPTPSSGNTLILSSATGAGTAPLSNGKK80 pKa = 9.21CLTPASSNGLTGFYY94 pKa = 10.76DD95 pKa = 3.67WTCATNFLLADD106 pKa = 4.11LQSAPTYY113 pKa = 10.02TGLFDD118 pKa = 4.03AYY120 pKa = 9.24KK121 pKa = 10.04IKK123 pKa = 10.47KK124 pKa = 7.24ITMNIEE130 pKa = 3.89YY131 pKa = 10.29LNNTSSVNSTGLMPTIYY148 pKa = 10.22AYY150 pKa = 10.61FDD152 pKa = 3.5QDD154 pKa = 3.34DD155 pKa = 4.01SAVPVSVADD164 pKa = 3.18ITRR167 pKa = 11.84RR168 pKa = 11.84QGVKK172 pKa = 10.12KK173 pKa = 10.04FQFGNKK179 pKa = 8.65SKK181 pKa = 9.89TSFSFSLKK189 pKa = 8.54PTVSSALEE197 pKa = 4.31TNSGTLIQAAVQKK210 pKa = 10.64AGWINCTQFAIPHH223 pKa = 5.99YY224 pKa = 8.57GLKK227 pKa = 10.44FAVTDD232 pKa = 4.58LYY234 pKa = 11.39LPGVNAIQQAFRR246 pKa = 11.84INWTYY251 pKa = 9.78VVSYY255 pKa = 10.21RR256 pKa = 11.84SPLRR260 pKa = 11.84NNN262 pKa = 3.27
Molecular weight: 28.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
488 |
226 |
262 |
244.0 |
27.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.197 ± 1.925 | 1.639 ± 0.382 |
4.098 ± 0.808 | 5.328 ± 3.528 |
3.893 ± 1.121 | 4.713 ± 1.077 |
1.025 ± 0.497 | 5.533 ± 0.441 |
6.352 ± 0.105 | 6.557 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.049 ± 0.109 | 5.943 ± 0.168 |
5.738 ± 1.17 | 6.148 ± 1.507 |
5.123 ± 0.125 | 7.377 ± 1.083 |
9.221 ± 0.838 | 5.943 ± 0.463 |
1.434 ± 0.224 | 3.689 ± 0.394 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
