
Microcella alkaliphila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microcella
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
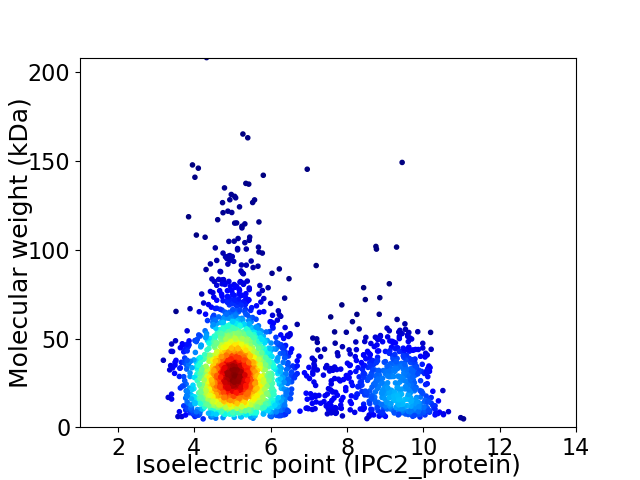
Virtual 2D-PAGE plot for 2621 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4WTU3|A0A0U4WTU3_9MICO Membrane protein OS=Microcella alkaliphila OX=279828 GN=MalAC0309_0414 PE=3 SV=1
MM1 pKa = 7.19KK2 pKa = 10.13HH3 pKa = 5.97SKK5 pKa = 8.45RR6 pKa = 11.84TRR8 pKa = 11.84AVLAVTAALSMTALAACAPVAEE30 pKa = 5.02PEE32 pKa = 4.39GQEE35 pKa = 3.63QTLNIAYY42 pKa = 8.53WNYY45 pKa = 10.17GPPAEE50 pKa = 5.11AGNEE54 pKa = 4.06AIANGFMEE62 pKa = 5.39ANPDD66 pKa = 3.57VKK68 pKa = 10.53VTLTPIAGDD77 pKa = 3.14NWGSYY82 pKa = 7.74YY83 pKa = 11.34ANLATLIASGEE94 pKa = 4.14RR95 pKa = 11.84PDD97 pKa = 4.36LAFTASEE104 pKa = 4.57GIKK107 pKa = 10.23FLSEE111 pKa = 3.77NGLVVPINEE120 pKa = 4.27YY121 pKa = 11.02LEE123 pKa = 4.6SDD125 pKa = 3.69PDD127 pKa = 3.43AAAIQDD133 pKa = 4.52DD134 pKa = 4.24IASEE138 pKa = 4.14LLDD141 pKa = 3.73SFAVEE146 pKa = 4.37GNITALPNGWNNMVVYY162 pKa = 10.62YY163 pKa = 8.79NTAVFDD169 pKa = 4.34AAGLPYY175 pKa = 10.17PEE177 pKa = 5.74SDD179 pKa = 3.36WTWEE183 pKa = 4.05EE184 pKa = 3.86FRR186 pKa = 11.84EE187 pKa = 4.08TAAALSADD195 pKa = 3.88TNGDD199 pKa = 3.42GVNDD203 pKa = 3.42QFGFTWASNEE213 pKa = 3.91IFPGILPWVANAGGNLVNDD232 pKa = 4.69DD233 pKa = 3.79VTEE236 pKa = 4.22ATANSAPVVEE246 pKa = 4.84AVSFLKK252 pKa = 11.0GLIDD256 pKa = 5.5DD257 pKa = 6.16GIAPAPSPMGDD268 pKa = 2.72IFTAFQNGNVAMFGAGRR285 pKa = 11.84WPSATFLPAGFTDD298 pKa = 3.69WDD300 pKa = 3.47IQIYY304 pKa = 7.38PTGATYY310 pKa = 8.34QTVAGANGYY319 pKa = 10.64AILTSAANPDD329 pKa = 4.16LAWEE333 pKa = 4.13LQKK336 pKa = 10.24YY337 pKa = 5.24TASAEE342 pKa = 4.25IQDD345 pKa = 4.26SLIGTPEE352 pKa = 3.99APRR355 pKa = 11.84DD356 pKa = 4.14AIPTTRR362 pKa = 11.84STAQKK367 pKa = 8.98TVDD370 pKa = 4.39AGIPPQNGALFYY382 pKa = 11.05GSVDD386 pKa = 3.95DD387 pKa = 5.16YY388 pKa = 11.42PVLSPFPAPAQYY400 pKa = 10.56SEE402 pKa = 4.61YY403 pKa = 9.64EE404 pKa = 3.98ATVLRR409 pKa = 11.84FVQLIFAGEE418 pKa = 4.12VAVEE422 pKa = 4.56DD423 pKa = 5.39GLDD426 pKa = 3.4QLQAEE431 pKa = 4.83LTALISGG438 pKa = 4.21
MM1 pKa = 7.19KK2 pKa = 10.13HH3 pKa = 5.97SKK5 pKa = 8.45RR6 pKa = 11.84TRR8 pKa = 11.84AVLAVTAALSMTALAACAPVAEE30 pKa = 5.02PEE32 pKa = 4.39GQEE35 pKa = 3.63QTLNIAYY42 pKa = 8.53WNYY45 pKa = 10.17GPPAEE50 pKa = 5.11AGNEE54 pKa = 4.06AIANGFMEE62 pKa = 5.39ANPDD66 pKa = 3.57VKK68 pKa = 10.53VTLTPIAGDD77 pKa = 3.14NWGSYY82 pKa = 7.74YY83 pKa = 11.34ANLATLIASGEE94 pKa = 4.14RR95 pKa = 11.84PDD97 pKa = 4.36LAFTASEE104 pKa = 4.57GIKK107 pKa = 10.23FLSEE111 pKa = 3.77NGLVVPINEE120 pKa = 4.27YY121 pKa = 11.02LEE123 pKa = 4.6SDD125 pKa = 3.69PDD127 pKa = 3.43AAAIQDD133 pKa = 4.52DD134 pKa = 4.24IASEE138 pKa = 4.14LLDD141 pKa = 3.73SFAVEE146 pKa = 4.37GNITALPNGWNNMVVYY162 pKa = 10.62YY163 pKa = 8.79NTAVFDD169 pKa = 4.34AAGLPYY175 pKa = 10.17PEE177 pKa = 5.74SDD179 pKa = 3.36WTWEE183 pKa = 4.05EE184 pKa = 3.86FRR186 pKa = 11.84EE187 pKa = 4.08TAAALSADD195 pKa = 3.88TNGDD199 pKa = 3.42GVNDD203 pKa = 3.42QFGFTWASNEE213 pKa = 3.91IFPGILPWVANAGGNLVNDD232 pKa = 4.69DD233 pKa = 3.79VTEE236 pKa = 4.22ATANSAPVVEE246 pKa = 4.84AVSFLKK252 pKa = 11.0GLIDD256 pKa = 5.5DD257 pKa = 6.16GIAPAPSPMGDD268 pKa = 2.72IFTAFQNGNVAMFGAGRR285 pKa = 11.84WPSATFLPAGFTDD298 pKa = 3.69WDD300 pKa = 3.47IQIYY304 pKa = 7.38PTGATYY310 pKa = 8.34QTVAGANGYY319 pKa = 10.64AILTSAANPDD329 pKa = 4.16LAWEE333 pKa = 4.13LQKK336 pKa = 10.24YY337 pKa = 5.24TASAEE342 pKa = 4.25IQDD345 pKa = 4.26SLIGTPEE352 pKa = 3.99APRR355 pKa = 11.84DD356 pKa = 4.14AIPTTRR362 pKa = 11.84STAQKK367 pKa = 8.98TVDD370 pKa = 4.39AGIPPQNGALFYY382 pKa = 11.05GSVDD386 pKa = 3.95DD387 pKa = 5.16YY388 pKa = 11.42PVLSPFPAPAQYY400 pKa = 10.56SEE402 pKa = 4.61YY403 pKa = 9.64EE404 pKa = 3.98ATVLRR409 pKa = 11.84FVQLIFAGEE418 pKa = 4.12VAVEE422 pKa = 4.56DD423 pKa = 5.39GLDD426 pKa = 3.4QLQAEE431 pKa = 4.83LTALISGG438 pKa = 4.21
Molecular weight: 46.34 kDa
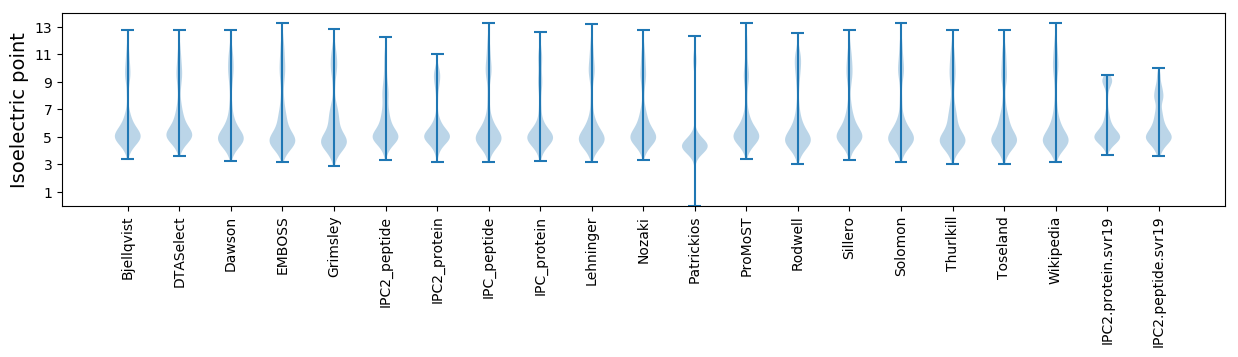
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4NX59|A0A0U4NX59_9MICO TVP38/TMEM64 family membrane protein OS=Microcella alkaliphila OX=279828 GN=MalAC0309_1958 PE=3 SV=1
MM1 pKa = 7.62KK2 pKa = 10.08VRR4 pKa = 11.84NSIKK8 pKa = 10.54SMKK11 pKa = 9.89KK12 pKa = 8.04MPGSQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84VFVINKK30 pKa = 7.43QNPRR34 pKa = 11.84FKK36 pKa = 10.8ARR38 pKa = 11.84QGG40 pKa = 3.28
MM1 pKa = 7.62KK2 pKa = 10.08VRR4 pKa = 11.84NSIKK8 pKa = 10.54SMKK11 pKa = 9.89KK12 pKa = 8.04MPGSQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84VFVINKK30 pKa = 7.43QNPRR34 pKa = 11.84FKK36 pKa = 10.8ARR38 pKa = 11.84QGG40 pKa = 3.28
Molecular weight: 4.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
817268 |
40 |
1977 |
311.8 |
33.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.662 ± 0.078 | 0.477 ± 0.01 |
6.394 ± 0.041 | 5.812 ± 0.041 |
2.988 ± 0.03 | 8.743 ± 0.043 |
2.049 ± 0.025 | 4.715 ± 0.036 |
1.855 ± 0.039 | 10.047 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.79 ± 0.019 | 1.97 ± 0.028 |
5.459 ± 0.039 | 2.724 ± 0.024 |
7.605 ± 0.051 | 5.359 ± 0.031 |
6.041 ± 0.035 | 8.996 ± 0.043 |
1.427 ± 0.02 | 1.886 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
