
bacterium 1XD8-76
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
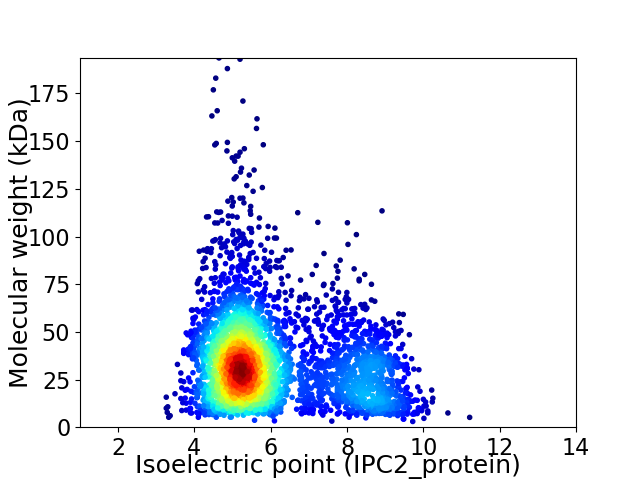
Virtual 2D-PAGE plot for 3768 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9EV71|A0A3A9EV71_9BACT Uncharacterized protein OS=bacterium 1XD8-76 OX=2320115 GN=D7X98_19615 PE=4 SV=1
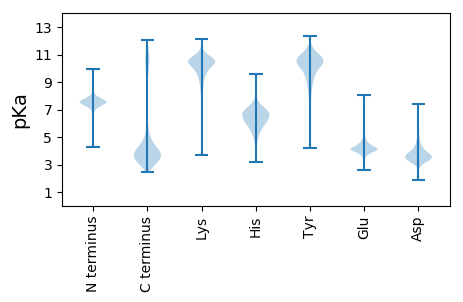
MM1 pKa = 7.61KK2 pKa = 10.44KK3 pKa = 10.24LFSVTLTGVVAFGLIACGNNAEE25 pKa = 4.18EE26 pKa = 4.68APAEE30 pKa = 4.04EE31 pKa = 4.45SSRR34 pKa = 11.84TEE36 pKa = 3.84SAEE39 pKa = 4.05EE40 pKa = 4.0PGSEE44 pKa = 4.08AEE46 pKa = 4.33EE47 pKa = 4.26PASEE51 pKa = 3.96EE52 pKa = 4.17AEE54 pKa = 4.19GGEE57 pKa = 4.35EE58 pKa = 4.93VTLEE62 pKa = 3.97WQQWWAVEE70 pKa = 4.0APEE73 pKa = 5.82GYY75 pKa = 8.62VQNMVDD81 pKa = 3.68KK82 pKa = 9.93YY83 pKa = 10.44YY84 pKa = 10.7EE85 pKa = 4.0EE86 pKa = 3.98TGVRR90 pKa = 11.84VEE92 pKa = 4.44LLSAPFADD100 pKa = 3.58TKK102 pKa = 8.88TQIVSGVSTGTVADD116 pKa = 3.63IVSVDD121 pKa = 3.58GSWVYY126 pKa = 11.45DD127 pKa = 3.81FADD130 pKa = 3.42QGVLTNMSTLMDD142 pKa = 4.62SIGFDD147 pKa = 3.04KK148 pKa = 10.87SLPASTWDD156 pKa = 3.79VNGSTYY162 pKa = 10.95AVPVLNFAYY171 pKa = 9.85PMFANQDD178 pKa = 3.38ILDD181 pKa = 4.08EE182 pKa = 4.85CGVTEE187 pKa = 5.73LPKK190 pKa = 9.41TWSDD194 pKa = 3.88FEE196 pKa = 4.44EE197 pKa = 4.68VCQKK201 pKa = 10.42IVDD204 pKa = 3.61KK205 pKa = 11.09GYY207 pKa = 10.91YY208 pKa = 9.57PYY210 pKa = 11.46ALNLDD215 pKa = 3.81TSSPSGIQNVYY226 pKa = 9.91MGFAWASDD234 pKa = 2.45IKK236 pKa = 10.63MKK238 pKa = 11.13NEE240 pKa = 4.34DD241 pKa = 3.55GNFDD245 pKa = 3.68VVNNPEE251 pKa = 4.08LKK253 pKa = 10.37EE254 pKa = 3.62YY255 pKa = 11.21AEE257 pKa = 4.67WIKK260 pKa = 11.09SLNDD264 pKa = 2.99KK265 pKa = 10.99GYY267 pKa = 9.19IYY269 pKa = 10.48PGMSSLTEE277 pKa = 3.52TDD279 pKa = 3.18MTSKK283 pKa = 10.51FSSGNICFIINSAAAITAWRR303 pKa = 11.84ADD305 pKa = 3.71APDD308 pKa = 4.34LNLTAAPIPVKK319 pKa = 10.76DD320 pKa = 4.23DD321 pKa = 3.48FTGEE325 pKa = 3.98SGMCVANWSLGITEE339 pKa = 4.11NSEE342 pKa = 4.13HH343 pKa = 7.26KK344 pKa = 10.44EE345 pKa = 3.6EE346 pKa = 3.94AMKK349 pKa = 10.71FIEE352 pKa = 4.19WLLNADD358 pKa = 4.45DD359 pKa = 5.38GGVCADD365 pKa = 4.34LAVTQTAFPNSTLAKK380 pKa = 9.16PDD382 pKa = 3.67YY383 pKa = 11.18SNGDD387 pKa = 3.44PVFQDD392 pKa = 3.19VYY394 pKa = 11.94AMYY397 pKa = 10.58QSGYY401 pKa = 9.17PYY403 pKa = 11.22NEE405 pKa = 3.63FTGMKK410 pKa = 8.8EE411 pKa = 3.99ANTIMTDD418 pKa = 3.78YY419 pKa = 10.72IDD421 pKa = 4.25EE422 pKa = 4.69LVTYY426 pKa = 9.55MDD428 pKa = 4.88GDD430 pKa = 3.93TDD432 pKa = 3.36VDD434 pKa = 3.75TFMSNLQEE442 pKa = 4.33TLDD445 pKa = 3.79EE446 pKa = 4.77VYY448 pKa = 11.08GKK450 pKa = 10.58
MM1 pKa = 7.61KK2 pKa = 10.44KK3 pKa = 10.24LFSVTLTGVVAFGLIACGNNAEE25 pKa = 4.18EE26 pKa = 4.68APAEE30 pKa = 4.04EE31 pKa = 4.45SSRR34 pKa = 11.84TEE36 pKa = 3.84SAEE39 pKa = 4.05EE40 pKa = 4.0PGSEE44 pKa = 4.08AEE46 pKa = 4.33EE47 pKa = 4.26PASEE51 pKa = 3.96EE52 pKa = 4.17AEE54 pKa = 4.19GGEE57 pKa = 4.35EE58 pKa = 4.93VTLEE62 pKa = 3.97WQQWWAVEE70 pKa = 4.0APEE73 pKa = 5.82GYY75 pKa = 8.62VQNMVDD81 pKa = 3.68KK82 pKa = 9.93YY83 pKa = 10.44YY84 pKa = 10.7EE85 pKa = 4.0EE86 pKa = 3.98TGVRR90 pKa = 11.84VEE92 pKa = 4.44LLSAPFADD100 pKa = 3.58TKK102 pKa = 8.88TQIVSGVSTGTVADD116 pKa = 3.63IVSVDD121 pKa = 3.58GSWVYY126 pKa = 11.45DD127 pKa = 3.81FADD130 pKa = 3.42QGVLTNMSTLMDD142 pKa = 4.62SIGFDD147 pKa = 3.04KK148 pKa = 10.87SLPASTWDD156 pKa = 3.79VNGSTYY162 pKa = 10.95AVPVLNFAYY171 pKa = 9.85PMFANQDD178 pKa = 3.38ILDD181 pKa = 4.08EE182 pKa = 4.85CGVTEE187 pKa = 5.73LPKK190 pKa = 9.41TWSDD194 pKa = 3.88FEE196 pKa = 4.44EE197 pKa = 4.68VCQKK201 pKa = 10.42IVDD204 pKa = 3.61KK205 pKa = 11.09GYY207 pKa = 10.91YY208 pKa = 9.57PYY210 pKa = 11.46ALNLDD215 pKa = 3.81TSSPSGIQNVYY226 pKa = 9.91MGFAWASDD234 pKa = 2.45IKK236 pKa = 10.63MKK238 pKa = 11.13NEE240 pKa = 4.34DD241 pKa = 3.55GNFDD245 pKa = 3.68VVNNPEE251 pKa = 4.08LKK253 pKa = 10.37EE254 pKa = 3.62YY255 pKa = 11.21AEE257 pKa = 4.67WIKK260 pKa = 11.09SLNDD264 pKa = 2.99KK265 pKa = 10.99GYY267 pKa = 9.19IYY269 pKa = 10.48PGMSSLTEE277 pKa = 3.52TDD279 pKa = 3.18MTSKK283 pKa = 10.51FSSGNICFIINSAAAITAWRR303 pKa = 11.84ADD305 pKa = 3.71APDD308 pKa = 4.34LNLTAAPIPVKK319 pKa = 10.76DD320 pKa = 4.23DD321 pKa = 3.48FTGEE325 pKa = 3.98SGMCVANWSLGITEE339 pKa = 4.11NSEE342 pKa = 4.13HH343 pKa = 7.26KK344 pKa = 10.44EE345 pKa = 3.6EE346 pKa = 3.94AMKK349 pKa = 10.71FIEE352 pKa = 4.19WLLNADD358 pKa = 4.45DD359 pKa = 5.38GGVCADD365 pKa = 4.34LAVTQTAFPNSTLAKK380 pKa = 9.16PDD382 pKa = 3.67YY383 pKa = 11.18SNGDD387 pKa = 3.44PVFQDD392 pKa = 3.19VYY394 pKa = 11.94AMYY397 pKa = 10.58QSGYY401 pKa = 9.17PYY403 pKa = 11.22NEE405 pKa = 3.63FTGMKK410 pKa = 8.8EE411 pKa = 3.99ANTIMTDD418 pKa = 3.78YY419 pKa = 10.72IDD421 pKa = 4.25EE422 pKa = 4.69LVTYY426 pKa = 9.55MDD428 pKa = 4.88GDD430 pKa = 3.93TDD432 pKa = 3.36VDD434 pKa = 3.75TFMSNLQEE442 pKa = 4.33TLDD445 pKa = 3.79EE446 pKa = 4.77VYY448 pKa = 11.08GKK450 pKa = 10.58
Molecular weight: 49.43 kDa
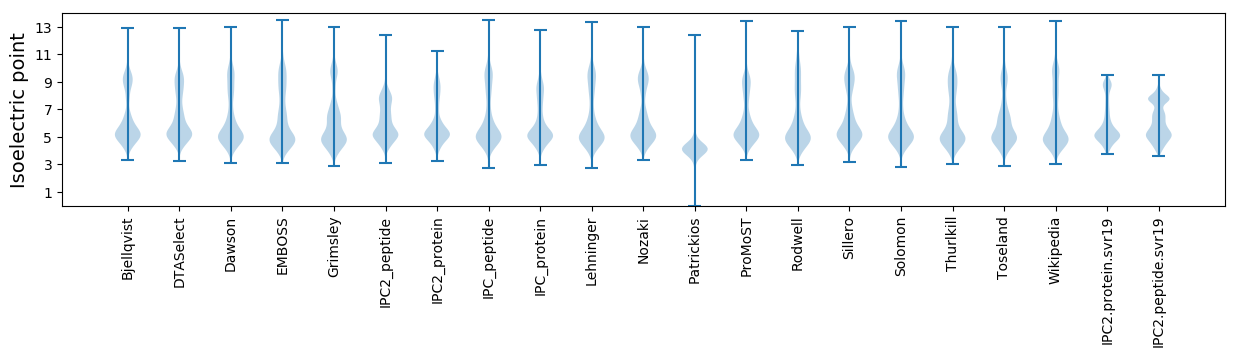
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9FKK9|A0A3A9FKK9_9BACT DeoR/GlpR transcriptional regulator OS=bacterium 1XD8-76 OX=2320115 GN=D7X98_01990 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.89RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.89RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1202740 |
26 |
1765 |
319.2 |
35.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.739 ± 0.042 | 1.586 ± 0.015 |
5.466 ± 0.03 | 8.318 ± 0.048 |
4.177 ± 0.031 | 7.424 ± 0.038 |
1.613 ± 0.014 | 7.034 ± 0.038 |
6.384 ± 0.038 | 9.052 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.183 ± 0.02 | 3.998 ± 0.028 |
3.246 ± 0.022 | 3.088 ± 0.02 |
5.122 ± 0.036 | 5.591 ± 0.031 |
4.978 ± 0.026 | 6.783 ± 0.033 |
0.996 ± 0.015 | 4.222 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
