
Faeces associated gemycircularvirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus geras1; Gerygone associated gemycircularvirus 1
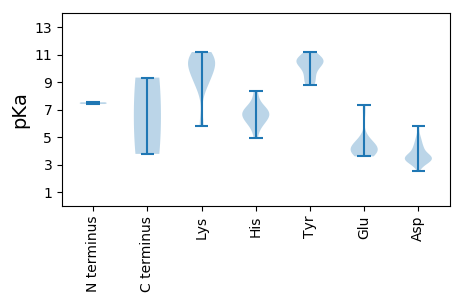
Average proteome isoelectric point is 7.42
Get precalculated fractions of proteins
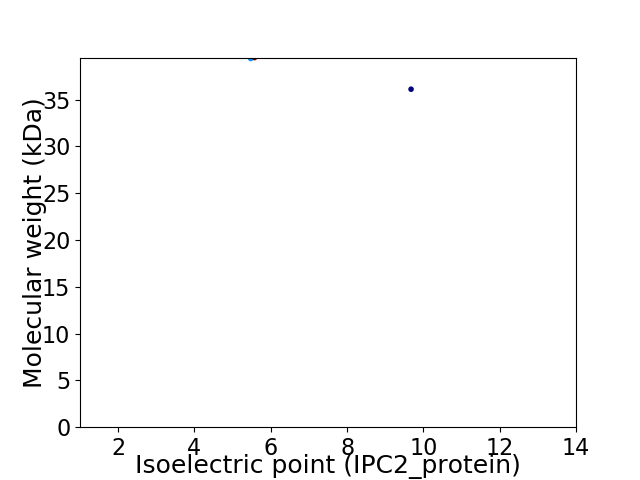
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YTB5|T1YTB5_9VIRU Replication-associated protein OS=Faeces associated gemycircularvirus 6 OX=1391035 PE=3 SV=1
MM1 pKa = 7.4LFVNSRR7 pKa = 11.84YY8 pKa = 10.26VLLTYY13 pKa = 9.15AQCGDD18 pKa = 4.04LDD20 pKa = 3.67PWSVSNHH27 pKa = 4.96LSALGAEE34 pKa = 4.68CIVARR39 pKa = 11.84EE40 pKa = 4.05IHH42 pKa = 6.37PTTGGIHH49 pKa = 5.94LHH51 pKa = 6.55CFADD55 pKa = 4.79FNRR58 pKa = 11.84KK59 pKa = 8.41FRR61 pKa = 11.84SRR63 pKa = 11.84SARR66 pKa = 11.84IFDD69 pKa = 3.35VDD71 pKa = 3.29GRR73 pKa = 11.84HH74 pKa = 6.11PNVVPSRR81 pKa = 11.84GTPEE85 pKa = 3.6KK86 pKa = 10.76GYY88 pKa = 10.89DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.86DD94 pKa = 3.32GDD96 pKa = 3.69VRR98 pKa = 11.84AGGLGRR104 pKa = 11.84PAPRR108 pKa = 11.84GGMSVGAHH116 pKa = 5.72ALVNVAHH123 pKa = 6.8LCEE126 pKa = 4.55DD127 pKa = 3.34TTEE130 pKa = 3.89FLEE133 pKa = 5.06LHH135 pKa = 7.02DD136 pKa = 4.25EE137 pKa = 4.15MDD139 pKa = 3.55RR140 pKa = 11.84SGLIARR146 pKa = 11.84FNNVRR151 pKa = 11.84AYY153 pKa = 10.56ADD155 pKa = 2.93WRR157 pKa = 11.84FRR159 pKa = 11.84PEE161 pKa = 3.76PVVYY165 pKa = 10.49ASPDD169 pKa = 3.4GVDD172 pKa = 4.02FRR174 pKa = 11.84SGSTDD179 pKa = 3.69GRR181 pKa = 11.84DD182 pKa = 3.16DD183 pKa = 3.47WLVQSRR189 pKa = 11.84IGDD192 pKa = 3.6EE193 pKa = 4.4LPFVSAPVANAPFAGYY209 pKa = 9.78IFCLLILYY217 pKa = 8.78GPSLTGKK224 pKa = 5.82TTWARR229 pKa = 11.84SLGDD233 pKa = 3.68HH234 pKa = 6.97IFIQGVLSGKK244 pKa = 9.67EE245 pKa = 3.84VLNSDD250 pKa = 3.22EE251 pKa = 4.26SARR254 pKa = 11.84YY255 pKa = 8.86AVLDD259 pKa = 4.81DD260 pKa = 3.6IRR262 pKa = 11.84GGLKK266 pKa = 10.03FFPAWKK272 pKa = 9.72DD273 pKa = 3.06WLGGQRR279 pKa = 11.84WISVKK284 pKa = 10.17QMYY287 pKa = 9.78RR288 pKa = 11.84DD289 pKa = 4.32PILLKK294 pKa = 10.04WGRR297 pKa = 11.84PCIWCANRR305 pKa = 11.84DD306 pKa = 3.32PRR308 pKa = 11.84ADD310 pKa = 2.98IRR312 pKa = 11.84RR313 pKa = 11.84SIDD316 pKa = 3.24KK317 pKa = 10.8DD318 pKa = 3.57DD319 pKa = 3.85GCFMEE324 pKa = 7.34DD325 pKa = 4.31DD326 pKa = 4.17MDD328 pKa = 4.65WINANCIFVYY338 pKa = 10.45VDD340 pKa = 3.23EE341 pKa = 4.72SLVTFRR347 pKa = 11.84ASTEE351 pKa = 3.77
MM1 pKa = 7.4LFVNSRR7 pKa = 11.84YY8 pKa = 10.26VLLTYY13 pKa = 9.15AQCGDD18 pKa = 4.04LDD20 pKa = 3.67PWSVSNHH27 pKa = 4.96LSALGAEE34 pKa = 4.68CIVARR39 pKa = 11.84EE40 pKa = 4.05IHH42 pKa = 6.37PTTGGIHH49 pKa = 5.94LHH51 pKa = 6.55CFADD55 pKa = 4.79FNRR58 pKa = 11.84KK59 pKa = 8.41FRR61 pKa = 11.84SRR63 pKa = 11.84SARR66 pKa = 11.84IFDD69 pKa = 3.35VDD71 pKa = 3.29GRR73 pKa = 11.84HH74 pKa = 6.11PNVVPSRR81 pKa = 11.84GTPEE85 pKa = 3.6KK86 pKa = 10.76GYY88 pKa = 10.89DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.86DD94 pKa = 3.32GDD96 pKa = 3.69VRR98 pKa = 11.84AGGLGRR104 pKa = 11.84PAPRR108 pKa = 11.84GGMSVGAHH116 pKa = 5.72ALVNVAHH123 pKa = 6.8LCEE126 pKa = 4.55DD127 pKa = 3.34TTEE130 pKa = 3.89FLEE133 pKa = 5.06LHH135 pKa = 7.02DD136 pKa = 4.25EE137 pKa = 4.15MDD139 pKa = 3.55RR140 pKa = 11.84SGLIARR146 pKa = 11.84FNNVRR151 pKa = 11.84AYY153 pKa = 10.56ADD155 pKa = 2.93WRR157 pKa = 11.84FRR159 pKa = 11.84PEE161 pKa = 3.76PVVYY165 pKa = 10.49ASPDD169 pKa = 3.4GVDD172 pKa = 4.02FRR174 pKa = 11.84SGSTDD179 pKa = 3.69GRR181 pKa = 11.84DD182 pKa = 3.16DD183 pKa = 3.47WLVQSRR189 pKa = 11.84IGDD192 pKa = 3.6EE193 pKa = 4.4LPFVSAPVANAPFAGYY209 pKa = 9.78IFCLLILYY217 pKa = 8.78GPSLTGKK224 pKa = 5.82TTWARR229 pKa = 11.84SLGDD233 pKa = 3.68HH234 pKa = 6.97IFIQGVLSGKK244 pKa = 9.67EE245 pKa = 3.84VLNSDD250 pKa = 3.22EE251 pKa = 4.26SARR254 pKa = 11.84YY255 pKa = 8.86AVLDD259 pKa = 4.81DD260 pKa = 3.6IRR262 pKa = 11.84GGLKK266 pKa = 10.03FFPAWKK272 pKa = 9.72DD273 pKa = 3.06WLGGQRR279 pKa = 11.84WISVKK284 pKa = 10.17QMYY287 pKa = 9.78RR288 pKa = 11.84DD289 pKa = 4.32PILLKK294 pKa = 10.04WGRR297 pKa = 11.84PCIWCANRR305 pKa = 11.84DD306 pKa = 3.32PRR308 pKa = 11.84ADD310 pKa = 2.98IRR312 pKa = 11.84RR313 pKa = 11.84SIDD316 pKa = 3.24KK317 pKa = 10.8DD318 pKa = 3.57DD319 pKa = 3.85GCFMEE324 pKa = 7.34DD325 pKa = 4.31DD326 pKa = 4.17MDD328 pKa = 4.65WINANCIFVYY338 pKa = 10.45VDD340 pKa = 3.23EE341 pKa = 4.72SLVTFRR347 pKa = 11.84ASTEE351 pKa = 3.77
Molecular weight: 39.38 kDa
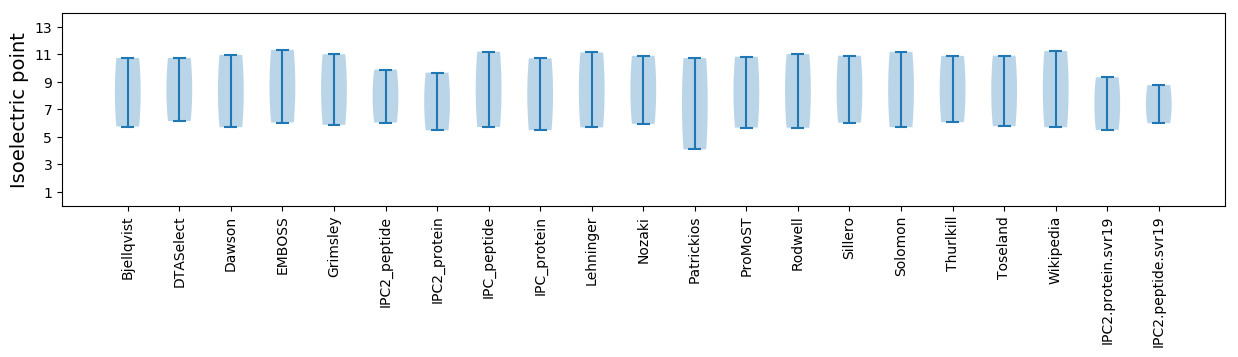
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YTB5|T1YTB5_9VIRU Replication-associated protein OS=Faeces associated gemycircularvirus 6 OX=1391035 PE=3 SV=1
MM1 pKa = 7.53PRR3 pKa = 11.84FARR6 pKa = 11.84RR7 pKa = 11.84SSRR10 pKa = 11.84FRR12 pKa = 11.84SRR14 pKa = 11.84KK15 pKa = 5.84TARR18 pKa = 11.84SSQRR22 pKa = 11.84TRR24 pKa = 11.84RR25 pKa = 11.84PARR28 pKa = 11.84RR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84TPRR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GMSTRR45 pKa = 11.84SILNKK50 pKa = 8.93TSRR53 pKa = 11.84KK54 pKa = 8.91KK55 pKa = 10.53RR56 pKa = 11.84NGMLTISNTQATGNPDD72 pKa = 3.44AFAQQPLTLIGSNTGGSIGVVHH94 pKa = 6.84FRR96 pKa = 11.84PTCMDD101 pKa = 4.27LASPTGVANSIVNQSQRR118 pKa = 11.84TSSTCFMRR126 pKa = 11.84GLAEE130 pKa = 3.9NLRR133 pKa = 11.84IEE135 pKa = 4.43TSSGNPWFHH144 pKa = 6.54RR145 pKa = 11.84RR146 pKa = 11.84VCITSRR152 pKa = 11.84DD153 pKa = 3.65DD154 pKa = 3.47NFRR157 pKa = 11.84LVATGDD163 pKa = 3.42NTGPQAAIMASGGIEE178 pKa = 4.11TSNGWQRR185 pKa = 11.84LAGNAQAGSEE195 pKa = 4.27LDD197 pKa = 3.32RR198 pKa = 11.84TIVVWLEE205 pKa = 3.59LLFKK209 pKa = 10.63GRR211 pKa = 11.84QGRR214 pKa = 11.84DD215 pKa = 2.54WDD217 pKa = 5.0DD218 pKa = 4.29IISAPVDD225 pKa = 3.42TTRR228 pKa = 11.84VDD230 pKa = 3.66LKK232 pKa = 10.88FDD234 pKa = 2.86KK235 pKa = 10.73TYY237 pKa = 10.4IYY239 pKa = 10.65RR240 pKa = 11.84SGNEE244 pKa = 3.69SGILKK249 pKa = 9.88EE250 pKa = 3.96KK251 pKa = 10.53KK252 pKa = 9.06LWHH255 pKa = 6.45RR256 pKa = 11.84MNKK259 pKa = 9.35NLYY262 pKa = 10.22FDD264 pKa = 5.5DD265 pKa = 5.82DD266 pKa = 3.91EE267 pKa = 6.7LGAGMEE273 pKa = 4.08SHH275 pKa = 7.61DD276 pKa = 4.56YY277 pKa = 11.19SVTDD281 pKa = 3.08KK282 pKa = 11.02RR283 pKa = 11.84GNGDD287 pKa = 3.09YY288 pKa = 10.74HH289 pKa = 8.32IFDD292 pKa = 4.99FFSQGSSGTTDD303 pKa = 3.4DD304 pKa = 3.63RR305 pKa = 11.84LKK307 pKa = 11.18VRR309 pKa = 11.84FTSTLYY315 pKa = 9.36WHH317 pKa = 7.08EE318 pKa = 4.2KK319 pKa = 9.32
MM1 pKa = 7.53PRR3 pKa = 11.84FARR6 pKa = 11.84RR7 pKa = 11.84SSRR10 pKa = 11.84FRR12 pKa = 11.84SRR14 pKa = 11.84KK15 pKa = 5.84TARR18 pKa = 11.84SSQRR22 pKa = 11.84TRR24 pKa = 11.84RR25 pKa = 11.84PARR28 pKa = 11.84RR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84TPRR35 pKa = 11.84RR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GMSTRR45 pKa = 11.84SILNKK50 pKa = 8.93TSRR53 pKa = 11.84KK54 pKa = 8.91KK55 pKa = 10.53RR56 pKa = 11.84NGMLTISNTQATGNPDD72 pKa = 3.44AFAQQPLTLIGSNTGGSIGVVHH94 pKa = 6.84FRR96 pKa = 11.84PTCMDD101 pKa = 4.27LASPTGVANSIVNQSQRR118 pKa = 11.84TSSTCFMRR126 pKa = 11.84GLAEE130 pKa = 3.9NLRR133 pKa = 11.84IEE135 pKa = 4.43TSSGNPWFHH144 pKa = 6.54RR145 pKa = 11.84RR146 pKa = 11.84VCITSRR152 pKa = 11.84DD153 pKa = 3.65DD154 pKa = 3.47NFRR157 pKa = 11.84LVATGDD163 pKa = 3.42NTGPQAAIMASGGIEE178 pKa = 4.11TSNGWQRR185 pKa = 11.84LAGNAQAGSEE195 pKa = 4.27LDD197 pKa = 3.32RR198 pKa = 11.84TIVVWLEE205 pKa = 3.59LLFKK209 pKa = 10.63GRR211 pKa = 11.84QGRR214 pKa = 11.84DD215 pKa = 2.54WDD217 pKa = 5.0DD218 pKa = 4.29IISAPVDD225 pKa = 3.42TTRR228 pKa = 11.84VDD230 pKa = 3.66LKK232 pKa = 10.88FDD234 pKa = 2.86KK235 pKa = 10.73TYY237 pKa = 10.4IYY239 pKa = 10.65RR240 pKa = 11.84SGNEE244 pKa = 3.69SGILKK249 pKa = 9.88EE250 pKa = 3.96KK251 pKa = 10.53KK252 pKa = 9.06LWHH255 pKa = 6.45RR256 pKa = 11.84MNKK259 pKa = 9.35NLYY262 pKa = 10.22FDD264 pKa = 5.5DD265 pKa = 5.82DD266 pKa = 3.91EE267 pKa = 6.7LGAGMEE273 pKa = 4.08SHH275 pKa = 7.61DD276 pKa = 4.56YY277 pKa = 11.19SVTDD281 pKa = 3.08KK282 pKa = 11.02RR283 pKa = 11.84GNGDD287 pKa = 3.09YY288 pKa = 10.74HH289 pKa = 8.32IFDD292 pKa = 4.99FFSQGSSGTTDD303 pKa = 3.4DD304 pKa = 3.63RR305 pKa = 11.84LKK307 pKa = 11.18VRR309 pKa = 11.84FTSTLYY315 pKa = 9.36WHH317 pKa = 7.08EE318 pKa = 4.2KK319 pKa = 9.32
Molecular weight: 36.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
670 |
319 |
351 |
335.0 |
37.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.866 ± 0.917 | 1.791 ± 0.638 |
8.06 ± 1.107 | 3.582 ± 0.335 |
4.776 ± 0.29 | 8.657 ± 0.145 |
2.239 ± 0.268 | 5.075 ± 0.279 |
3.582 ± 0.605 | 7.164 ± 0.671 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.314 | 4.328 ± 0.75 |
4.179 ± 0.783 | 2.388 ± 0.795 |
10.448 ± 1.568 | 8.358 ± 1.489 |
5.97 ± 2.105 | 5.522 ± 1.32 |
2.388 ± 0.38 | 2.537 ± 0.492 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
