
Pacific flying fox faeces associated circular DNA virus-3
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 9.04
Get precalculated fractions of proteins
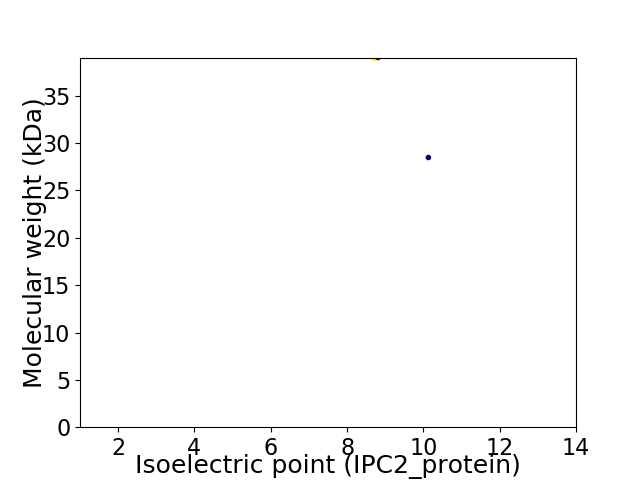
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
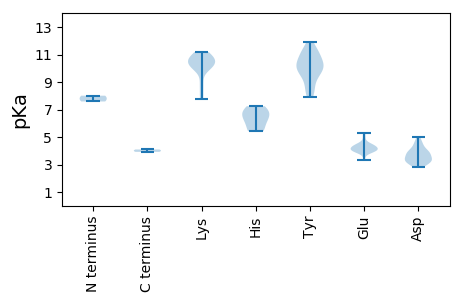
>tr|A0A140CTT1|A0A140CTT1_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-3 OX=1796012 PE=4 SV=1
MM1 pKa = 8.01PKK3 pKa = 10.22RR4 pKa = 11.84IRR6 pKa = 11.84QPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WVFTLNNFTEE22 pKa = 4.46EE23 pKa = 4.06EE24 pKa = 4.24LTTIKK29 pKa = 10.92NILTDD34 pKa = 4.24LAPTYY39 pKa = 10.6AIIGKK44 pKa = 8.81EE45 pKa = 3.95VGEE48 pKa = 4.93CGTPHH53 pKa = 6.16LQGFVSLKK61 pKa = 9.95KK62 pKa = 10.37QISFTAMKK70 pKa = 10.4SLIGTRR76 pKa = 11.84AHH78 pKa = 6.77IEE80 pKa = 3.95AANGTDD86 pKa = 3.33VDD88 pKa = 3.91NQLYY92 pKa = 10.28CSKK95 pKa = 10.6QDD97 pKa = 3.13ADD99 pKa = 3.24AWQLGEE105 pKa = 4.24PRR107 pKa = 11.84QSVTGRR113 pKa = 11.84GGNALDD119 pKa = 3.59YY120 pKa = 10.95RR121 pKa = 11.84GVLEE125 pKa = 4.4ALQSGSTVTEE135 pKa = 3.86IVKK138 pKa = 10.72NPDD141 pKa = 3.1LVVPLIIQHH150 pKa = 6.23RR151 pKa = 11.84NLRR154 pKa = 11.84QVLLDD159 pKa = 3.71LKK161 pKa = 10.85KK162 pKa = 10.76EE163 pKa = 4.01KK164 pKa = 10.27EE165 pKa = 4.14LEE167 pKa = 4.13KK168 pKa = 11.08LRR170 pKa = 11.84TAYY173 pKa = 10.0KK174 pKa = 10.5NVTLRR179 pKa = 11.84PFQEE183 pKa = 3.95EE184 pKa = 4.36LVNILSSDD192 pKa = 3.52PDD194 pKa = 3.32SRR196 pKa = 11.84AVYY199 pKa = 9.21WYY201 pKa = 9.78YY202 pKa = 11.12DD203 pKa = 3.25QYY205 pKa = 11.9GNTGKK210 pKa = 7.83TWFSKK215 pKa = 10.71YY216 pKa = 9.92LVARR220 pKa = 11.84MDD222 pKa = 3.78ALRR225 pKa = 11.84LEE227 pKa = 4.41NGKK230 pKa = 7.79TADD233 pKa = 3.98LKK235 pKa = 10.89HH236 pKa = 6.98AYY238 pKa = 8.69EE239 pKa = 4.26GQKK242 pKa = 9.96IVIFDD247 pKa = 3.88FSRR250 pKa = 11.84STMEE254 pKa = 4.49RR255 pKa = 11.84INYY258 pKa = 8.31EE259 pKa = 4.05CIEE262 pKa = 4.48AIKK265 pKa = 10.77NGLVFSGKK273 pKa = 10.13YY274 pKa = 9.66EE275 pKa = 4.09STSKK279 pKa = 10.31IFPVPHH285 pKa = 5.47VVCFANCPPDD295 pKa = 3.79TSKK298 pKa = 11.18LSYY301 pKa = 10.38DD302 pKa = 3.13RR303 pKa = 11.84WRR305 pKa = 11.84IRR307 pKa = 11.84CIDD310 pKa = 4.26EE311 pKa = 3.97INISVGGTYY320 pKa = 8.22QMRR323 pKa = 11.84WVRR326 pKa = 11.84GGSRR330 pKa = 11.84LRR332 pKa = 11.84RR333 pKa = 11.84SRR335 pKa = 11.84PAFLII340 pKa = 4.09
MM1 pKa = 8.01PKK3 pKa = 10.22RR4 pKa = 11.84IRR6 pKa = 11.84QPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WVFTLNNFTEE22 pKa = 4.46EE23 pKa = 4.06EE24 pKa = 4.24LTTIKK29 pKa = 10.92NILTDD34 pKa = 4.24LAPTYY39 pKa = 10.6AIIGKK44 pKa = 8.81EE45 pKa = 3.95VGEE48 pKa = 4.93CGTPHH53 pKa = 6.16LQGFVSLKK61 pKa = 9.95KK62 pKa = 10.37QISFTAMKK70 pKa = 10.4SLIGTRR76 pKa = 11.84AHH78 pKa = 6.77IEE80 pKa = 3.95AANGTDD86 pKa = 3.33VDD88 pKa = 3.91NQLYY92 pKa = 10.28CSKK95 pKa = 10.6QDD97 pKa = 3.13ADD99 pKa = 3.24AWQLGEE105 pKa = 4.24PRR107 pKa = 11.84QSVTGRR113 pKa = 11.84GGNALDD119 pKa = 3.59YY120 pKa = 10.95RR121 pKa = 11.84GVLEE125 pKa = 4.4ALQSGSTVTEE135 pKa = 3.86IVKK138 pKa = 10.72NPDD141 pKa = 3.1LVVPLIIQHH150 pKa = 6.23RR151 pKa = 11.84NLRR154 pKa = 11.84QVLLDD159 pKa = 3.71LKK161 pKa = 10.85KK162 pKa = 10.76EE163 pKa = 4.01KK164 pKa = 10.27EE165 pKa = 4.14LEE167 pKa = 4.13KK168 pKa = 11.08LRR170 pKa = 11.84TAYY173 pKa = 10.0KK174 pKa = 10.5NVTLRR179 pKa = 11.84PFQEE183 pKa = 3.95EE184 pKa = 4.36LVNILSSDD192 pKa = 3.52PDD194 pKa = 3.32SRR196 pKa = 11.84AVYY199 pKa = 9.21WYY201 pKa = 9.78YY202 pKa = 11.12DD203 pKa = 3.25QYY205 pKa = 11.9GNTGKK210 pKa = 7.83TWFSKK215 pKa = 10.71YY216 pKa = 9.92LVARR220 pKa = 11.84MDD222 pKa = 3.78ALRR225 pKa = 11.84LEE227 pKa = 4.41NGKK230 pKa = 7.79TADD233 pKa = 3.98LKK235 pKa = 10.89HH236 pKa = 6.98AYY238 pKa = 8.69EE239 pKa = 4.26GQKK242 pKa = 9.96IVIFDD247 pKa = 3.88FSRR250 pKa = 11.84STMEE254 pKa = 4.49RR255 pKa = 11.84INYY258 pKa = 8.31EE259 pKa = 4.05CIEE262 pKa = 4.48AIKK265 pKa = 10.77NGLVFSGKK273 pKa = 10.13YY274 pKa = 9.66EE275 pKa = 4.09STSKK279 pKa = 10.31IFPVPHH285 pKa = 5.47VVCFANCPPDD295 pKa = 3.79TSKK298 pKa = 11.18LSYY301 pKa = 10.38DD302 pKa = 3.13RR303 pKa = 11.84WRR305 pKa = 11.84IRR307 pKa = 11.84CIDD310 pKa = 4.26EE311 pKa = 3.97INISVGGTYY320 pKa = 8.22QMRR323 pKa = 11.84WVRR326 pKa = 11.84GGSRR330 pKa = 11.84LRR332 pKa = 11.84RR333 pKa = 11.84SRR335 pKa = 11.84PAFLII340 pKa = 4.09
Molecular weight: 38.94 kDa
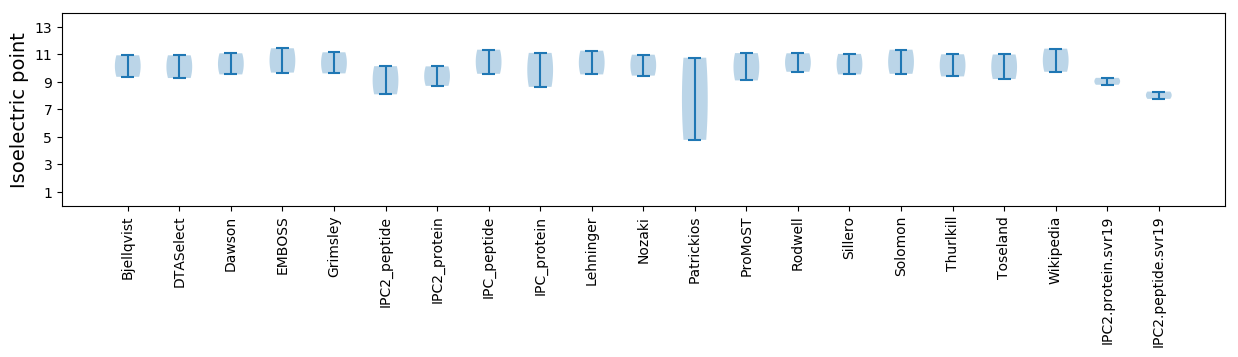
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTT1|A0A140CTT1_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-3 OX=1796012 PE=4 SV=1
MM1 pKa = 7.59VKK3 pKa = 10.13RR4 pKa = 11.84QSTLASWRR12 pKa = 11.84RR13 pKa = 11.84VKK15 pKa = 10.19RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84MVRR21 pKa = 11.84SRR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84SRR28 pKa = 11.84GRR30 pKa = 11.84SVLYY34 pKa = 9.77RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TFRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84FVRR47 pKa = 11.84RR48 pKa = 11.84NKK50 pKa = 9.95YY51 pKa = 10.36LSGIYY56 pKa = 10.22AQFTTPIKK64 pKa = 10.24WSDD67 pKa = 3.01IGDD70 pKa = 3.95DD71 pKa = 4.49GFATMNLNLNIAAMIGDD88 pKa = 4.55IQPQLTGYY96 pKa = 10.27RR97 pKa = 11.84VVFDD101 pKa = 3.37QAKK104 pKa = 8.43VLRR107 pKa = 11.84IKK109 pKa = 10.12RR110 pKa = 11.84TVWLNDD116 pKa = 2.81SDD118 pKa = 5.0EE119 pKa = 4.23YY120 pKa = 11.21TKK122 pKa = 11.1ADD124 pKa = 3.43QRR126 pKa = 11.84VTEE129 pKa = 5.27LYY131 pKa = 10.65SSYY134 pKa = 11.47DD135 pKa = 3.25PDD137 pKa = 4.69ANGSSLTQAQIIQDD151 pKa = 3.59PSHH154 pKa = 7.24RR155 pKa = 11.84MVIMKK160 pKa = 9.1PFSRR164 pKa = 11.84YY165 pKa = 9.65RR166 pKa = 11.84YY167 pKa = 7.94WIKK170 pKa = 10.68PNWRR174 pKa = 11.84LQNATQGTPALFSYY188 pKa = 9.64TGYY191 pKa = 11.27NDD193 pKa = 3.37ILDD196 pKa = 3.86WQADD200 pKa = 4.0RR201 pKa = 11.84TGNSANSVHH210 pKa = 5.43MVARR214 pKa = 11.84GTTEE218 pKa = 3.36QGAINYY224 pKa = 8.93QDD226 pKa = 2.91TYY228 pKa = 11.14YY229 pKa = 10.35IHH231 pKa = 6.62FKK233 pKa = 10.84KK234 pKa = 10.46RR235 pKa = 11.84IQNAGGTT242 pKa = 3.92
MM1 pKa = 7.59VKK3 pKa = 10.13RR4 pKa = 11.84QSTLASWRR12 pKa = 11.84RR13 pKa = 11.84VKK15 pKa = 10.19RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84MVRR21 pKa = 11.84SRR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84SRR28 pKa = 11.84GRR30 pKa = 11.84SVLYY34 pKa = 9.77RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TFRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84FVRR47 pKa = 11.84RR48 pKa = 11.84NKK50 pKa = 9.95YY51 pKa = 10.36LSGIYY56 pKa = 10.22AQFTTPIKK64 pKa = 10.24WSDD67 pKa = 3.01IGDD70 pKa = 3.95DD71 pKa = 4.49GFATMNLNLNIAAMIGDD88 pKa = 4.55IQPQLTGYY96 pKa = 10.27RR97 pKa = 11.84VVFDD101 pKa = 3.37QAKK104 pKa = 8.43VLRR107 pKa = 11.84IKK109 pKa = 10.12RR110 pKa = 11.84TVWLNDD116 pKa = 2.81SDD118 pKa = 5.0EE119 pKa = 4.23YY120 pKa = 11.21TKK122 pKa = 11.1ADD124 pKa = 3.43QRR126 pKa = 11.84VTEE129 pKa = 5.27LYY131 pKa = 10.65SSYY134 pKa = 11.47DD135 pKa = 3.25PDD137 pKa = 4.69ANGSSLTQAQIIQDD151 pKa = 3.59PSHH154 pKa = 7.24RR155 pKa = 11.84MVIMKK160 pKa = 9.1PFSRR164 pKa = 11.84YY165 pKa = 9.65RR166 pKa = 11.84YY167 pKa = 7.94WIKK170 pKa = 10.68PNWRR174 pKa = 11.84LQNATQGTPALFSYY188 pKa = 9.64TGYY191 pKa = 11.27NDD193 pKa = 3.37ILDD196 pKa = 3.86WQADD200 pKa = 4.0RR201 pKa = 11.84TGNSANSVHH210 pKa = 5.43MVARR214 pKa = 11.84GTTEE218 pKa = 3.36QGAINYY224 pKa = 8.93QDD226 pKa = 2.91TYY228 pKa = 11.14YY229 pKa = 10.35IHH231 pKa = 6.62FKK233 pKa = 10.84KK234 pKa = 10.46RR235 pKa = 11.84IQNAGGTT242 pKa = 3.92
Molecular weight: 28.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
582 |
242 |
340 |
291.0 |
33.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.014 ± 0.364 | 1.031 ± 0.628 |
5.498 ± 0.427 | 4.124 ± 1.758 |
3.436 ± 0.08 | 6.186 ± 0.244 |
1.375 ± 0.082 | 6.529 ± 0.202 |
5.67 ± 0.685 | 7.56 ± 1.334 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.062 ± 0.506 | 4.983 ± 0.237 |
3.608 ± 0.436 | 4.983 ± 0.741 |
10.481 ± 1.923 | 6.529 ± 0.302 |
6.873 ± 0.344 | 6.186 ± 0.244 |
2.062 ± 0.254 | 4.811 ± 0.594 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
