
Actinobacteria bacterium SCGC AG-212-D09
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; unclassified Actinobacteria
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
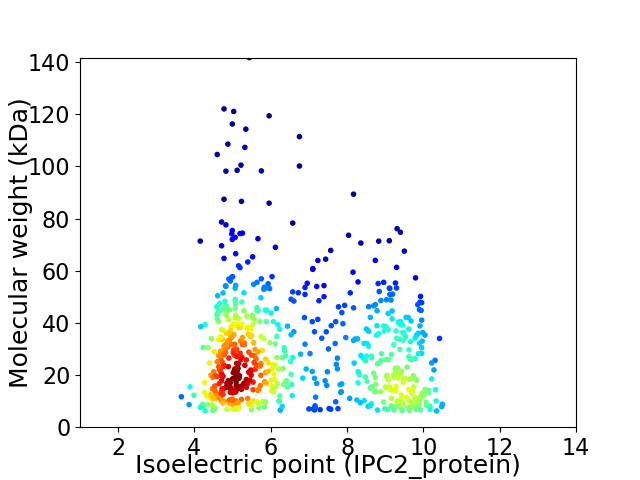
Virtual 2D-PAGE plot for 651 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
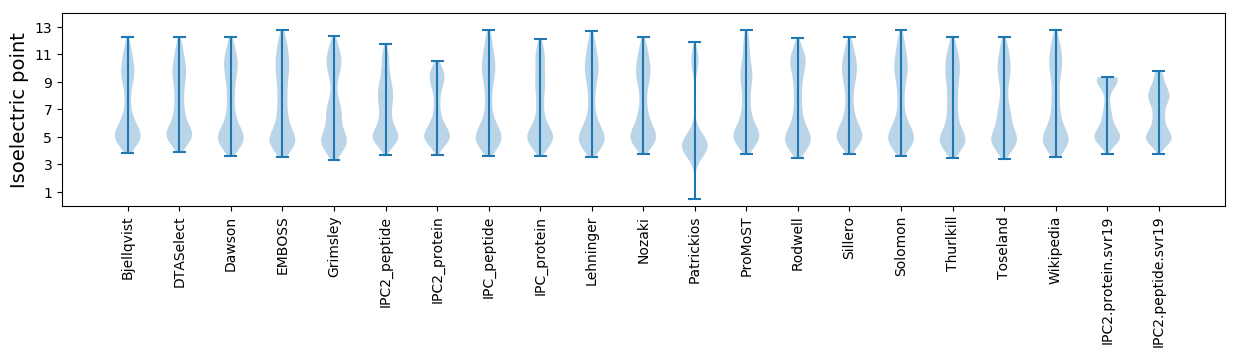
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177PYJ2|A0A177PYJ2_9ACTN Uncharacterized protein OS=Actinobacteria bacterium SCGC AG-212-D09 OX=1799652 GN=AYO39_01600 PE=4 SV=1
MM1 pKa = 6.85STEE4 pKa = 3.78ALFINVEE11 pKa = 3.84VDD13 pKa = 3.08EE14 pKa = 5.08SVANDD19 pKa = 3.49PALAAKK25 pKa = 8.55LTEE28 pKa = 4.13VCPVDD33 pKa = 5.16IFAQSDD39 pKa = 3.58DD40 pKa = 3.42GTLRR44 pKa = 11.84IVEE47 pKa = 4.43EE48 pKa = 4.12NLDD51 pKa = 3.79EE52 pKa = 5.13CVLCEE57 pKa = 4.18LCINAAPTGTVRR69 pKa = 11.84VIKK72 pKa = 10.2LYY74 pKa = 10.92SGEE77 pKa = 4.18TLPAA81 pKa = 4.79
MM1 pKa = 6.85STEE4 pKa = 3.78ALFINVEE11 pKa = 3.84VDD13 pKa = 3.08EE14 pKa = 5.08SVANDD19 pKa = 3.49PALAAKK25 pKa = 8.55LTEE28 pKa = 4.13VCPVDD33 pKa = 5.16IFAQSDD39 pKa = 3.58DD40 pKa = 3.42GTLRR44 pKa = 11.84IVEE47 pKa = 4.43EE48 pKa = 4.12NLDD51 pKa = 3.79EE52 pKa = 5.13CVLCEE57 pKa = 4.18LCINAAPTGTVRR69 pKa = 11.84VIKK72 pKa = 10.2LYY74 pKa = 10.92SGEE77 pKa = 4.18TLPAA81 pKa = 4.79
Molecular weight: 8.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177PXF9|A0A177PXF9_9ACTN PNPLA domain-containing protein OS=Actinobacteria bacterium SCGC AG-212-D09 OX=1799652 GN=AYO39_03130 PE=4 SV=1
MM1 pKa = 7.64QMIVIGVDD9 pKa = 2.89THH11 pKa = 6.4KK12 pKa = 10.69RR13 pKa = 11.84SHH15 pKa = 6.0TLVALDD21 pKa = 4.34AGTGGARR28 pKa = 11.84GQLTIAASADD38 pKa = 3.38GTLEE42 pKa = 3.85ALRR45 pKa = 11.84FAAALDD51 pKa = 3.95SEE53 pKa = 4.87RR54 pKa = 11.84VWAVEE59 pKa = 3.94DD60 pKa = 3.73CRR62 pKa = 11.84HH63 pKa = 5.0VSGRR67 pKa = 11.84LEE69 pKa = 4.13RR70 pKa = 11.84EE71 pKa = 4.02LLSSGDD77 pKa = 3.21RR78 pKa = 11.84VVRR81 pKa = 11.84VAPGLTEE88 pKa = 3.72NSRR91 pKa = 11.84RR92 pKa = 11.84AARR95 pKa = 11.84QPGKK99 pKa = 10.24SDD101 pKa = 4.87PIDD104 pKa = 3.41ATAIARR110 pKa = 11.84AALRR114 pKa = 11.84EE115 pKa = 4.68GIDD118 pKa = 3.51TLPVAFLDD126 pKa = 4.29EE127 pKa = 4.17QAHH130 pKa = 6.45EE131 pKa = 4.04IRR133 pKa = 11.84VLNDD137 pKa = 2.7YY138 pKa = 10.45RR139 pKa = 11.84RR140 pKa = 11.84QLVAEE145 pKa = 4.52RR146 pKa = 11.84VRR148 pKa = 11.84LINRR152 pKa = 11.84LRR154 pKa = 11.84WHH156 pKa = 7.05LVQIAPEE163 pKa = 3.9IEE165 pKa = 4.16AQIRR169 pKa = 11.84PAGLIGPRR177 pKa = 11.84IRR179 pKa = 11.84AKK181 pKa = 8.54VTRR184 pKa = 11.84QLARR188 pKa = 11.84LPHH191 pKa = 5.84SPQLRR196 pKa = 11.84VAKK199 pKa = 10.45AILKK203 pKa = 8.81RR204 pKa = 11.84VCEE207 pKa = 4.14NYY209 pKa = 9.96RR210 pKa = 11.84EE211 pKa = 4.14EE212 pKa = 4.33NEE214 pKa = 4.26LLIEE218 pKa = 4.61LKK220 pKa = 9.96TLIDD224 pKa = 3.4AHH226 pKa = 6.79CPQLLNQRR234 pKa = 11.84GCGTVTAAIIIGHH247 pKa = 5.06TAGAKK252 pKa = 9.75RR253 pKa = 11.84FPTDD257 pKa = 2.44ACFARR262 pKa = 11.84HH263 pKa = 6.1TGTAPIPASSGNTQRR278 pKa = 11.84HH279 pKa = 4.86RR280 pKa = 11.84LHH282 pKa = 7.08RR283 pKa = 11.84GGDD286 pKa = 3.27RR287 pKa = 11.84QLNHH291 pKa = 7.19ALHH294 pKa = 6.98IIALSRR300 pKa = 11.84ARR302 pKa = 11.84TDD304 pKa = 3.39PATRR308 pKa = 11.84AYY310 pKa = 10.83LDD312 pKa = 3.43RR313 pKa = 11.84RR314 pKa = 11.84HH315 pKa = 6.14TEE317 pKa = 3.59GKK319 pKa = 7.83TKK321 pKa = 10.63KK322 pKa = 9.71EE323 pKa = 3.65ALRR326 pKa = 11.84CLKK329 pKa = 10.25RR330 pKa = 11.84HH331 pKa = 5.57LARR334 pKa = 11.84RR335 pKa = 11.84IWRR338 pKa = 11.84LLYY341 pKa = 8.23TTIEE345 pKa = 4.13AAPPRR350 pKa = 11.84TDD352 pKa = 3.19PSLEE356 pKa = 3.81KK357 pKa = 10.85SEE359 pKa = 4.55IPNFTT364 pKa = 3.53
MM1 pKa = 7.64QMIVIGVDD9 pKa = 2.89THH11 pKa = 6.4KK12 pKa = 10.69RR13 pKa = 11.84SHH15 pKa = 6.0TLVALDD21 pKa = 4.34AGTGGARR28 pKa = 11.84GQLTIAASADD38 pKa = 3.38GTLEE42 pKa = 3.85ALRR45 pKa = 11.84FAAALDD51 pKa = 3.95SEE53 pKa = 4.87RR54 pKa = 11.84VWAVEE59 pKa = 3.94DD60 pKa = 3.73CRR62 pKa = 11.84HH63 pKa = 5.0VSGRR67 pKa = 11.84LEE69 pKa = 4.13RR70 pKa = 11.84EE71 pKa = 4.02LLSSGDD77 pKa = 3.21RR78 pKa = 11.84VVRR81 pKa = 11.84VAPGLTEE88 pKa = 3.72NSRR91 pKa = 11.84RR92 pKa = 11.84AARR95 pKa = 11.84QPGKK99 pKa = 10.24SDD101 pKa = 4.87PIDD104 pKa = 3.41ATAIARR110 pKa = 11.84AALRR114 pKa = 11.84EE115 pKa = 4.68GIDD118 pKa = 3.51TLPVAFLDD126 pKa = 4.29EE127 pKa = 4.17QAHH130 pKa = 6.45EE131 pKa = 4.04IRR133 pKa = 11.84VLNDD137 pKa = 2.7YY138 pKa = 10.45RR139 pKa = 11.84RR140 pKa = 11.84QLVAEE145 pKa = 4.52RR146 pKa = 11.84VRR148 pKa = 11.84LINRR152 pKa = 11.84LRR154 pKa = 11.84WHH156 pKa = 7.05LVQIAPEE163 pKa = 3.9IEE165 pKa = 4.16AQIRR169 pKa = 11.84PAGLIGPRR177 pKa = 11.84IRR179 pKa = 11.84AKK181 pKa = 8.54VTRR184 pKa = 11.84QLARR188 pKa = 11.84LPHH191 pKa = 5.84SPQLRR196 pKa = 11.84VAKK199 pKa = 10.45AILKK203 pKa = 8.81RR204 pKa = 11.84VCEE207 pKa = 4.14NYY209 pKa = 9.96RR210 pKa = 11.84EE211 pKa = 4.14EE212 pKa = 4.33NEE214 pKa = 4.26LLIEE218 pKa = 4.61LKK220 pKa = 9.96TLIDD224 pKa = 3.4AHH226 pKa = 6.79CPQLLNQRR234 pKa = 11.84GCGTVTAAIIIGHH247 pKa = 5.06TAGAKK252 pKa = 9.75RR253 pKa = 11.84FPTDD257 pKa = 2.44ACFARR262 pKa = 11.84HH263 pKa = 6.1TGTAPIPASSGNTQRR278 pKa = 11.84HH279 pKa = 4.86RR280 pKa = 11.84LHH282 pKa = 7.08RR283 pKa = 11.84GGDD286 pKa = 3.27RR287 pKa = 11.84QLNHH291 pKa = 7.19ALHH294 pKa = 6.98IIALSRR300 pKa = 11.84ARR302 pKa = 11.84TDD304 pKa = 3.39PATRR308 pKa = 11.84AYY310 pKa = 10.83LDD312 pKa = 3.43RR313 pKa = 11.84RR314 pKa = 11.84HH315 pKa = 6.14TEE317 pKa = 3.59GKK319 pKa = 7.83TKK321 pKa = 10.63KK322 pKa = 9.71EE323 pKa = 3.65ALRR326 pKa = 11.84CLKK329 pKa = 10.25RR330 pKa = 11.84HH331 pKa = 5.57LARR334 pKa = 11.84RR335 pKa = 11.84IWRR338 pKa = 11.84LLYY341 pKa = 8.23TTIEE345 pKa = 4.13AAPPRR350 pKa = 11.84TDD352 pKa = 3.19PSLEE356 pKa = 3.81KK357 pKa = 10.85SEE359 pKa = 4.55IPNFTT364 pKa = 3.53
Molecular weight: 40.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
184414 |
59 |
1282 |
283.3 |
30.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.857 ± 0.139 | 0.876 ± 0.032 |
5.533 ± 0.084 | 5.622 ± 0.129 |
3.088 ± 0.053 | 9.095 ± 0.13 |
2.152 ± 0.045 | 4.167 ± 0.064 |
2.108 ± 0.068 | 9.994 ± 0.134 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.662 ± 0.04 | 2.317 ± 0.066 |
5.882 ± 0.077 | 3.165 ± 0.058 |
8.122 ± 0.129 | 6.114 ± 0.092 |
5.818 ± 0.108 | 7.837 ± 0.081 |
1.436 ± 0.037 | 2.155 ± 0.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
