
Streptomyces niveus NCIMB 11891
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces niveus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
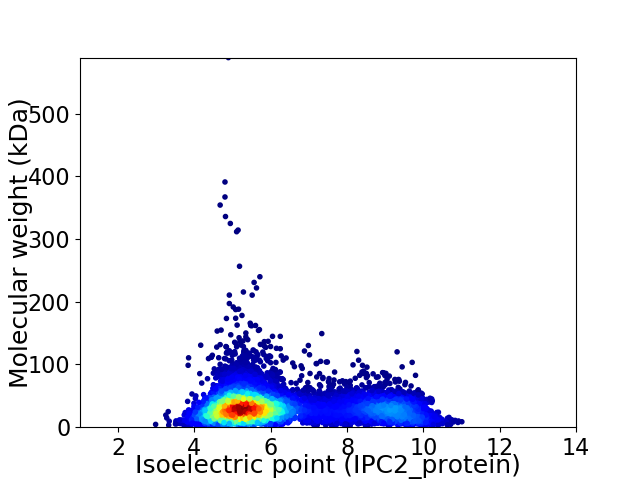
Virtual 2D-PAGE plot for 7746 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
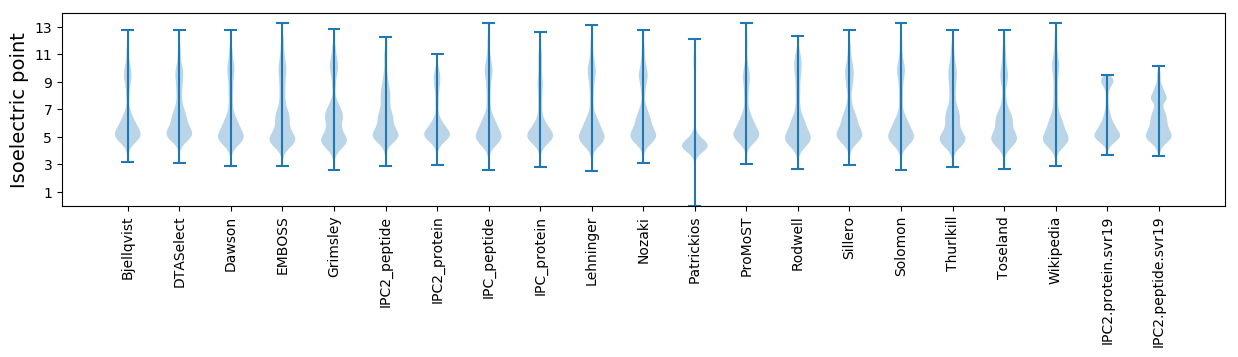
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V6KJK0|V6KJK0_STRNV OmpA-like domain-containing protein OS=Streptomyces niveus NCIMB 11891 OX=1352941 GN=M877_06490 PE=4 SV=1
MM1 pKa = 7.34LTTRR5 pKa = 11.84YY6 pKa = 10.08VPGSPNWADD15 pKa = 3.5LATPDD20 pKa = 4.18IEE22 pKa = 4.61GAKK25 pKa = 9.0TFYY28 pKa = 10.89GGVLGWDD35 pKa = 3.67HH36 pKa = 6.64VPAGPEE42 pKa = 3.6FGGYY46 pKa = 11.01GMFQLNGKK54 pKa = 6.64TVAGVMQLPAEE65 pKa = 4.28TVPLSWTLYY74 pKa = 9.88FQTADD79 pKa = 3.21ATATSSAVKK88 pKa = 10.59DD89 pKa = 3.51NGGTVVNEE97 pKa = 3.93PMDD100 pKa = 3.98VEE102 pKa = 4.27DD103 pKa = 4.6LGRR106 pKa = 11.84MTLCADD112 pKa = 3.51PAGAGFATWQPGTNTGLEE130 pKa = 4.18ATDD133 pKa = 4.17TPGTLCWAEE142 pKa = 4.85LYY144 pKa = 10.39TPDD147 pKa = 3.02VDD149 pKa = 4.35AAVGFYY155 pKa = 10.91GSVFGWEE162 pKa = 4.29TTTMPMPGGGGDD174 pKa = 3.55YY175 pKa = 11.46VMVHH179 pKa = 6.55PAGKK183 pKa = 10.57GPDD186 pKa = 3.3GMFAGVVEE194 pKa = 4.2NRR196 pKa = 11.84PDD198 pKa = 3.35TGLGATGPNWLLYY211 pKa = 10.52FGTADD216 pKa = 3.75CDD218 pKa = 3.69AAVAASDD225 pKa = 3.71RR226 pKa = 11.84LGGTTVQEE234 pKa = 4.08PTDD237 pKa = 3.52IEE239 pKa = 4.59GVGRR243 pKa = 11.84FAVLTDD249 pKa = 3.86PYY251 pKa = 10.32GARR254 pKa = 11.84LAVMQGVSQDD264 pKa = 3.21TT265 pKa = 3.52
MM1 pKa = 7.34LTTRR5 pKa = 11.84YY6 pKa = 10.08VPGSPNWADD15 pKa = 3.5LATPDD20 pKa = 4.18IEE22 pKa = 4.61GAKK25 pKa = 9.0TFYY28 pKa = 10.89GGVLGWDD35 pKa = 3.67HH36 pKa = 6.64VPAGPEE42 pKa = 3.6FGGYY46 pKa = 11.01GMFQLNGKK54 pKa = 6.64TVAGVMQLPAEE65 pKa = 4.28TVPLSWTLYY74 pKa = 9.88FQTADD79 pKa = 3.21ATATSSAVKK88 pKa = 10.59DD89 pKa = 3.51NGGTVVNEE97 pKa = 3.93PMDD100 pKa = 3.98VEE102 pKa = 4.27DD103 pKa = 4.6LGRR106 pKa = 11.84MTLCADD112 pKa = 3.51PAGAGFATWQPGTNTGLEE130 pKa = 4.18ATDD133 pKa = 4.17TPGTLCWAEE142 pKa = 4.85LYY144 pKa = 10.39TPDD147 pKa = 3.02VDD149 pKa = 4.35AAVGFYY155 pKa = 10.91GSVFGWEE162 pKa = 4.29TTTMPMPGGGGDD174 pKa = 3.55YY175 pKa = 11.46VMVHH179 pKa = 6.55PAGKK183 pKa = 10.57GPDD186 pKa = 3.3GMFAGVVEE194 pKa = 4.2NRR196 pKa = 11.84PDD198 pKa = 3.35TGLGATGPNWLLYY211 pKa = 10.52FGTADD216 pKa = 3.75CDD218 pKa = 3.69AAVAASDD225 pKa = 3.71RR226 pKa = 11.84LGGTTVQEE234 pKa = 4.08PTDD237 pKa = 3.52IEE239 pKa = 4.59GVGRR243 pKa = 11.84FAVLTDD249 pKa = 3.86PYY251 pKa = 10.32GARR254 pKa = 11.84LAVMQGVSQDD264 pKa = 3.21TT265 pKa = 3.52
Molecular weight: 27.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V6K2B0|V6K2B0_STRNV Membrane protein OS=Streptomyces niveus NCIMB 11891 OX=1352941 GN=M877_20115 PE=4 SV=1
MM1 pKa = 7.49HH2 pKa = 7.21SGVIASFSWAHH13 pKa = 6.57GDD15 pKa = 3.54SQAMSRR21 pKa = 11.84LPNHH25 pKa = 6.51HH26 pKa = 6.67LWSPRR31 pKa = 11.84VSKK34 pKa = 10.08RR35 pKa = 11.84TFQPNNRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84AKK46 pKa = 8.7THH48 pKa = 5.15GFRR51 pKa = 11.84LRR53 pKa = 11.84MRR55 pKa = 11.84TRR57 pKa = 11.84AGRR60 pKa = 11.84AILANRR66 pKa = 11.84RR67 pKa = 11.84GKK69 pKa = 10.51GRR71 pKa = 11.84ANLSAA76 pKa = 4.66
MM1 pKa = 7.49HH2 pKa = 7.21SGVIASFSWAHH13 pKa = 6.57GDD15 pKa = 3.54SQAMSRR21 pKa = 11.84LPNHH25 pKa = 6.51HH26 pKa = 6.67LWSPRR31 pKa = 11.84VSKK34 pKa = 10.08RR35 pKa = 11.84TFQPNNRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84AKK46 pKa = 8.7THH48 pKa = 5.15GFRR51 pKa = 11.84LRR53 pKa = 11.84MRR55 pKa = 11.84TRR57 pKa = 11.84AGRR60 pKa = 11.84AILANRR66 pKa = 11.84RR67 pKa = 11.84GKK69 pKa = 10.51GRR71 pKa = 11.84ANLSAA76 pKa = 4.66
Molecular weight: 8.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2460520 |
27 |
5582 |
317.7 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.364 ± 0.038 | 0.731 ± 0.007 |
6.159 ± 0.025 | 5.66 ± 0.025 |
2.736 ± 0.017 | 9.542 ± 0.028 |
2.232 ± 0.015 | 3.263 ± 0.019 |
2.215 ± 0.025 | 10.305 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.75 ± 0.011 | 1.852 ± 0.017 |
6.046 ± 0.024 | 2.686 ± 0.016 |
7.915 ± 0.038 | 5.229 ± 0.019 |
6.297 ± 0.026 | 8.436 ± 0.03 |
1.501 ± 0.013 | 2.082 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
