
Duck adenovirus 1
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Atadenovirus; Duck atadenovirus A
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
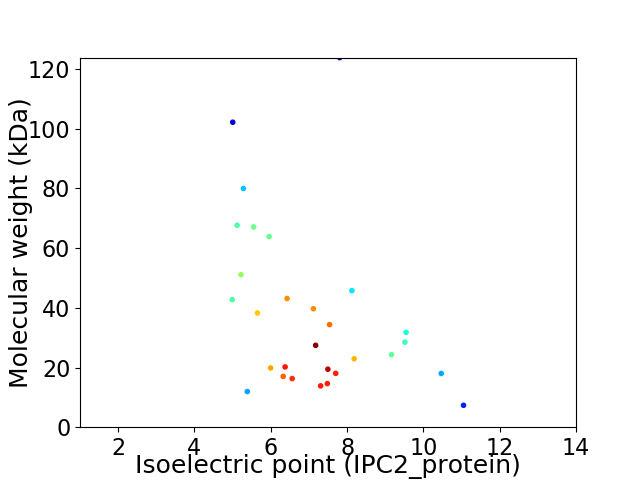
Virtual 2D-PAGE plot for 29 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
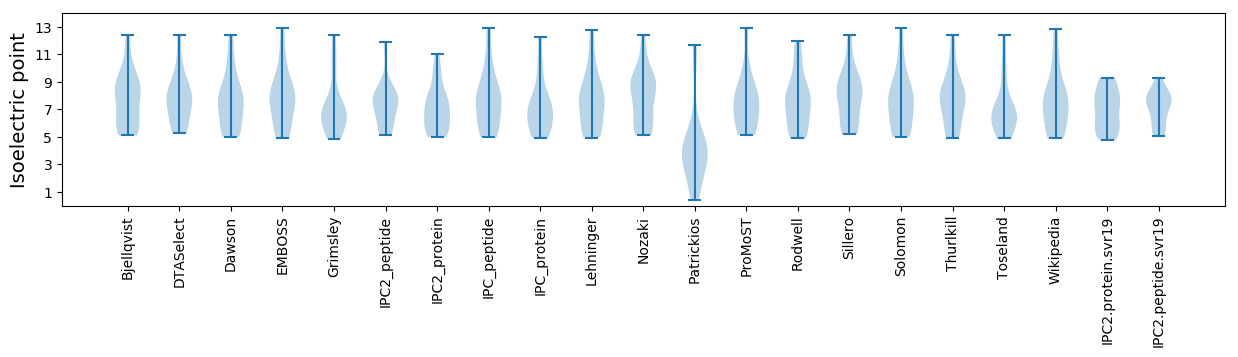
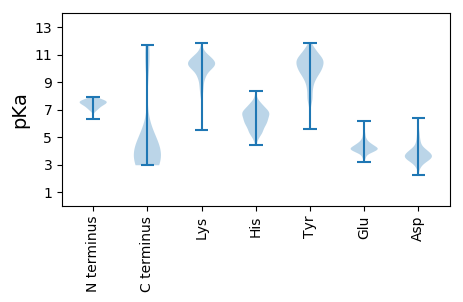
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O11413|O11413_9ADEN IVa2 protein OS=Duck adenovirus 1 OX=130329 GN=IVa2 PE=4 SV=1
MM1 pKa = 7.93AFRR4 pKa = 11.84PAPPPAPFTAFPYY17 pKa = 9.6LTGNLPTWSVSTVKK31 pKa = 10.44NDD33 pKa = 3.2PSMPLGAYY41 pKa = 10.57LMLPSEE47 pKa = 4.26NPEE50 pKa = 4.05DD51 pKa = 3.91VFNKK55 pKa = 9.52YY56 pKa = 7.0QTVYY60 pKa = 10.6LVPGEE65 pKa = 4.07QYY67 pKa = 9.77RR68 pKa = 11.84WKK70 pKa = 10.47QVIIKK75 pKa = 10.33SSITIHH81 pKa = 5.84GQGALVKK88 pKa = 10.25LDD90 pKa = 3.71GPGPSLTISGGSGLPIDD107 pKa = 4.05MTVEE111 pKa = 3.56LRR113 pKa = 11.84DD114 pKa = 3.65INFKK118 pKa = 11.02GLDD121 pKa = 3.37IPPDD125 pKa = 3.63RR126 pKa = 11.84EE127 pKa = 4.07EE128 pKa = 4.25LMEE131 pKa = 4.79DD132 pKa = 2.78AFLRR136 pKa = 11.84HH137 pKa = 5.37SAIWSHH143 pKa = 5.2NVTRR147 pKa = 11.84TVINGCSFVNFKK159 pKa = 10.38GAAVWFYY166 pKa = 11.53DD167 pKa = 5.14EE168 pKa = 4.44DD169 pKa = 4.39TTWSGRR175 pKa = 11.84IFAEE179 pKa = 3.37QHH181 pKa = 6.02LFINNRR187 pKa = 11.84ITGCRR192 pKa = 11.84IGLALGSVSRR202 pKa = 11.84NSVASHH208 pKa = 6.85NIFNDD213 pKa = 3.15CHH215 pKa = 7.03VCFNVLGGGWTLVNNCIMNSRR236 pKa = 11.84CAYY239 pKa = 9.33LHH241 pKa = 6.52AKK243 pKa = 9.21EE244 pKa = 4.34GMWYY248 pKa = 10.16SGPATSGPALVNVFSGNVLNNCAANGNMWPTDD280 pKa = 3.64YY281 pKa = 11.23LGGGDD286 pKa = 4.25VPLKK290 pKa = 10.56LAAFYY295 pKa = 10.7FDD297 pKa = 5.81DD298 pKa = 4.66EE299 pKa = 4.64DD300 pKa = 5.56AIPPVWTGNAHH311 pKa = 5.38NWADD315 pKa = 3.42VTLTKK320 pKa = 10.55FSSDD324 pKa = 3.33LEE326 pKa = 4.6SYY328 pKa = 10.86SITGCMFIGQTTAVPSAGRR347 pKa = 11.84IYY349 pKa = 10.83VGNTISDD356 pKa = 3.2KK357 pKa = 11.26VYY359 pKa = 10.61FFGCSGNGVYY369 pKa = 10.58LYY371 pKa = 9.14GTTDD375 pKa = 3.39ANLFPDD381 pKa = 4.81DD382 pKa = 4.5FGTAQSGNPPSS393 pKa = 3.43
MM1 pKa = 7.93AFRR4 pKa = 11.84PAPPPAPFTAFPYY17 pKa = 9.6LTGNLPTWSVSTVKK31 pKa = 10.44NDD33 pKa = 3.2PSMPLGAYY41 pKa = 10.57LMLPSEE47 pKa = 4.26NPEE50 pKa = 4.05DD51 pKa = 3.91VFNKK55 pKa = 9.52YY56 pKa = 7.0QTVYY60 pKa = 10.6LVPGEE65 pKa = 4.07QYY67 pKa = 9.77RR68 pKa = 11.84WKK70 pKa = 10.47QVIIKK75 pKa = 10.33SSITIHH81 pKa = 5.84GQGALVKK88 pKa = 10.25LDD90 pKa = 3.71GPGPSLTISGGSGLPIDD107 pKa = 4.05MTVEE111 pKa = 3.56LRR113 pKa = 11.84DD114 pKa = 3.65INFKK118 pKa = 11.02GLDD121 pKa = 3.37IPPDD125 pKa = 3.63RR126 pKa = 11.84EE127 pKa = 4.07EE128 pKa = 4.25LMEE131 pKa = 4.79DD132 pKa = 2.78AFLRR136 pKa = 11.84HH137 pKa = 5.37SAIWSHH143 pKa = 5.2NVTRR147 pKa = 11.84TVINGCSFVNFKK159 pKa = 10.38GAAVWFYY166 pKa = 11.53DD167 pKa = 5.14EE168 pKa = 4.44DD169 pKa = 4.39TTWSGRR175 pKa = 11.84IFAEE179 pKa = 3.37QHH181 pKa = 6.02LFINNRR187 pKa = 11.84ITGCRR192 pKa = 11.84IGLALGSVSRR202 pKa = 11.84NSVASHH208 pKa = 6.85NIFNDD213 pKa = 3.15CHH215 pKa = 7.03VCFNVLGGGWTLVNNCIMNSRR236 pKa = 11.84CAYY239 pKa = 9.33LHH241 pKa = 6.52AKK243 pKa = 9.21EE244 pKa = 4.34GMWYY248 pKa = 10.16SGPATSGPALVNVFSGNVLNNCAANGNMWPTDD280 pKa = 3.64YY281 pKa = 11.23LGGGDD286 pKa = 4.25VPLKK290 pKa = 10.56LAAFYY295 pKa = 10.7FDD297 pKa = 5.81DD298 pKa = 4.66EE299 pKa = 4.64DD300 pKa = 5.56AIPPVWTGNAHH311 pKa = 5.38NWADD315 pKa = 3.42VTLTKK320 pKa = 10.55FSSDD324 pKa = 3.33LEE326 pKa = 4.6SYY328 pKa = 10.86SITGCMFIGQTTAVPSAGRR347 pKa = 11.84IYY349 pKa = 10.83VGNTISDD356 pKa = 3.2KK357 pKa = 11.26VYY359 pKa = 10.61FFGCSGNGVYY369 pKa = 10.58LYY371 pKa = 9.14GTTDD375 pKa = 3.39ANLFPDD381 pKa = 4.81DD382 pKa = 4.5FGTAQSGNPPSS393 pKa = 3.43
Molecular weight: 42.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O11420|O11420_9ADEN PVI protein OS=Duck adenovirus 1 OX=130329 GN=pVI PE=4 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84RR3 pKa = 11.84SRR5 pKa = 11.84SYY7 pKa = 9.91GGLRR11 pKa = 11.84YY12 pKa = 8.69GHH14 pKa = 5.63SVVRR18 pKa = 11.84YY19 pKa = 8.22RR20 pKa = 11.84RR21 pKa = 11.84SSQVRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84LKK35 pKa = 10.99GGFLPAIIPLIAAAISAAPAIAGTVIAAKK64 pKa = 10.16NARR67 pKa = 3.7
MM1 pKa = 7.46RR2 pKa = 11.84RR3 pKa = 11.84SRR5 pKa = 11.84SYY7 pKa = 9.91GGLRR11 pKa = 11.84YY12 pKa = 8.69GHH14 pKa = 5.63SVVRR18 pKa = 11.84YY19 pKa = 8.22RR20 pKa = 11.84RR21 pKa = 11.84SSQVRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84PRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84LKK35 pKa = 10.99GGFLPAIIPLIAAAISAAPAIAGTVIAAKK64 pKa = 10.16NARR67 pKa = 3.7
Molecular weight: 7.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9891 |
67 |
1079 |
341.1 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.451 ± 0.497 | 2.396 ± 0.43 |
4.802 ± 0.345 | 5.136 ± 0.396 |
4.459 ± 0.255 | 5.732 ± 0.549 |
2.204 ± 0.216 | 5.49 ± 0.288 |
4.671 ± 0.557 | 9.837 ± 0.396 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.022 ± 0.179 | 5.348 ± 0.403 |
6.036 ± 0.302 | 3.923 ± 0.278 |
5.763 ± 0.599 | 6.541 ± 0.235 |
6.855 ± 0.452 | 6.066 ± 0.376 |
1.415 ± 0.15 | 3.852 ± 0.302 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
