
Neorhizobium sp. NCHU2750
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Neorhizobium; unclassified Neorhizobium
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
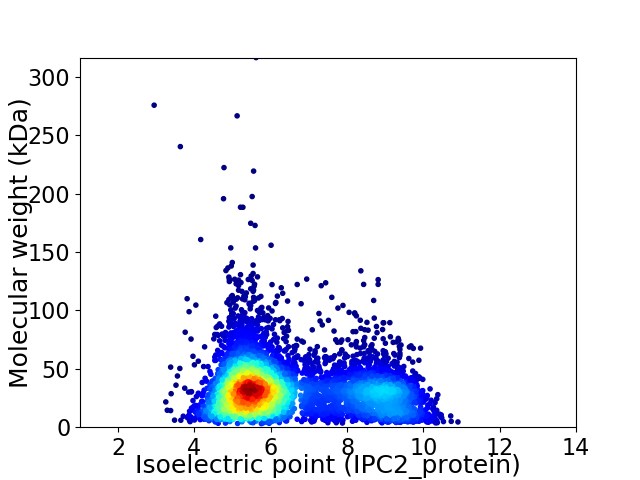
Virtual 2D-PAGE plot for 5901 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A386DVE1|A0A386DVE1_9RHIZ Amino acid ABC transporter permease OS=Neorhizobium sp. NCHU2750 OX=1825976 GN=NCHU2750_12160 PE=3 SV=1
MM1 pKa = 7.43LLKK4 pKa = 10.77SKK6 pKa = 9.26VLCLLASSALVGIVSLASTANASVGSDD33 pKa = 2.87ILGLSSDD40 pKa = 3.67GSVMLNYY47 pKa = 10.48SYY49 pKa = 11.41DD50 pKa = 3.68ADD52 pKa = 4.39TEE54 pKa = 4.43TGTYY58 pKa = 10.98LLTTTSTGEE67 pKa = 4.19TQQITPPAGAMSVYY81 pKa = 10.13PSALSADD88 pKa = 3.78GKK90 pKa = 9.01TVTGYY95 pKa = 10.99YY96 pKa = 8.71VTDD99 pKa = 4.57AYY101 pKa = 11.21QKK103 pKa = 10.88VAFVWTAAGGFVDD116 pKa = 5.88LGVLDD121 pKa = 4.93EE122 pKa = 4.75DD123 pKa = 4.15TTGAFAQSVSGDD135 pKa = 3.25GSAVAGYY142 pKa = 10.35VSIDD146 pKa = 3.03SYY148 pKa = 10.33MAEE151 pKa = 3.8GFYY154 pKa = 10.27WSSSTGMVGIGTLGGRR170 pKa = 11.84DD171 pKa = 3.31SYY173 pKa = 12.25ANAISTDD180 pKa = 2.74GSTVVGRR187 pKa = 11.84ADD189 pKa = 3.23TADD192 pKa = 3.53EE193 pKa = 4.26VFHH196 pKa = 6.74AFVWSVDD203 pKa = 3.44TNTMQNIDD211 pKa = 3.65TLYY214 pKa = 8.72TSSSATLVSDD224 pKa = 4.94DD225 pKa = 3.52GTAVAGTGSTGTTSGVFRR243 pKa = 11.84WTADD247 pKa = 2.66TGMVDD252 pKa = 2.79IGTLGGIYY260 pKa = 9.93TSVAAMSGDD269 pKa = 3.47GNVLVGQSTTVGEE282 pKa = 4.47SDD284 pKa = 3.42YY285 pKa = 11.24HH286 pKa = 7.22AYY288 pKa = 10.22RR289 pKa = 11.84YY290 pKa = 8.91VASTSTMTDD299 pKa = 2.84LGTLGGTYY307 pKa = 9.98SYY309 pKa = 11.63ASDD312 pKa = 3.64LTADD316 pKa = 3.71GSIVVGRR323 pKa = 11.84ASDD326 pKa = 3.52SASAYY331 pKa = 10.43HH332 pKa = 6.04GFVWSEE338 pKa = 3.74ATGMQTVEE346 pKa = 4.31DD347 pKa = 4.18WLTAHH352 pKa = 7.4GATLEE357 pKa = 4.05GDD359 pKa = 3.61MTTSADD365 pKa = 3.99KK366 pKa = 10.72ISADD370 pKa = 3.07GSVIVGSTTSGSTYY384 pKa = 9.18IARR387 pKa = 11.84VVTTGSDD394 pKa = 3.36DD395 pKa = 3.61STGGDD400 pKa = 3.09SGGGDD405 pKa = 3.8SEE407 pKa = 5.29CNADD411 pKa = 3.47VCDD414 pKa = 4.27GGGEE418 pKa = 4.09SGIIDD423 pKa = 3.6TAKK426 pKa = 10.52YY427 pKa = 9.65FPTVASANNMIVQSGVNSADD447 pKa = 3.14TIMFGAQGAPMRR459 pKa = 11.84NLLKK463 pKa = 10.05TGQKK467 pKa = 10.0SVWGTVDD474 pKa = 3.41GGYY477 pKa = 10.55DD478 pKa = 4.11DD479 pKa = 5.82SDD481 pKa = 3.91HH482 pKa = 7.27ADD484 pKa = 3.36GGLALGDD491 pKa = 3.66FGFGYY496 pKa = 10.79GIAEE500 pKa = 4.16GVTARR505 pKa = 11.84FSAGMTYY512 pKa = 10.19TDD514 pKa = 3.33QDD516 pKa = 3.6LDD518 pKa = 3.46AGGDD522 pKa = 3.34VRR524 pKa = 11.84QRR526 pKa = 11.84GFYY529 pKa = 10.58LSPEE533 pKa = 3.99VSADD537 pKa = 3.29LGRR540 pKa = 11.84NVYY543 pKa = 7.73MTVGGYY549 pKa = 8.24WGRR552 pKa = 11.84SSIDD556 pKa = 2.95SRR558 pKa = 11.84RR559 pKa = 11.84GYY561 pKa = 11.27ASGAITDD568 pKa = 3.68YY569 pKa = 11.38SYY571 pKa = 12.1GDD573 pKa = 3.67TNAEE577 pKa = 3.61TWGAKK582 pKa = 9.1IRR584 pKa = 11.84FDD586 pKa = 3.61WLNAATIADD595 pKa = 4.1TAITPYY601 pKa = 10.89AGLSYY606 pKa = 11.16AHH608 pKa = 6.19TRR610 pKa = 11.84VDD612 pKa = 3.56GFSEE616 pKa = 4.14TGGAFPVEE624 pKa = 4.21FDD626 pKa = 3.74GSSDD630 pKa = 3.08HH631 pKa = 6.65ATIARR636 pKa = 11.84LGSDD640 pKa = 3.85FVRR643 pKa = 11.84PLNDD647 pKa = 3.37TVRR650 pKa = 11.84LLAKK654 pKa = 10.58AEE656 pKa = 3.94LDD658 pKa = 3.76YY659 pKa = 11.43QFEE662 pKa = 4.15NHH664 pKa = 6.24AAATTGTLTGISDD677 pKa = 4.48FDD679 pKa = 5.42LEE681 pKa = 4.54GQDD684 pKa = 4.99LQQFWVRR691 pKa = 11.84GGLGAEE697 pKa = 3.87FDD699 pKa = 3.79VGKK702 pKa = 10.65GVASFMVNATTKK714 pKa = 10.62GQDD717 pKa = 3.08PTVWLRR723 pKa = 11.84SNYY726 pKa = 7.13TVKK729 pKa = 10.76FF730 pKa = 3.7
MM1 pKa = 7.43LLKK4 pKa = 10.77SKK6 pKa = 9.26VLCLLASSALVGIVSLASTANASVGSDD33 pKa = 2.87ILGLSSDD40 pKa = 3.67GSVMLNYY47 pKa = 10.48SYY49 pKa = 11.41DD50 pKa = 3.68ADD52 pKa = 4.39TEE54 pKa = 4.43TGTYY58 pKa = 10.98LLTTTSTGEE67 pKa = 4.19TQQITPPAGAMSVYY81 pKa = 10.13PSALSADD88 pKa = 3.78GKK90 pKa = 9.01TVTGYY95 pKa = 10.99YY96 pKa = 8.71VTDD99 pKa = 4.57AYY101 pKa = 11.21QKK103 pKa = 10.88VAFVWTAAGGFVDD116 pKa = 5.88LGVLDD121 pKa = 4.93EE122 pKa = 4.75DD123 pKa = 4.15TTGAFAQSVSGDD135 pKa = 3.25GSAVAGYY142 pKa = 10.35VSIDD146 pKa = 3.03SYY148 pKa = 10.33MAEE151 pKa = 3.8GFYY154 pKa = 10.27WSSSTGMVGIGTLGGRR170 pKa = 11.84DD171 pKa = 3.31SYY173 pKa = 12.25ANAISTDD180 pKa = 2.74GSTVVGRR187 pKa = 11.84ADD189 pKa = 3.23TADD192 pKa = 3.53EE193 pKa = 4.26VFHH196 pKa = 6.74AFVWSVDD203 pKa = 3.44TNTMQNIDD211 pKa = 3.65TLYY214 pKa = 8.72TSSSATLVSDD224 pKa = 4.94DD225 pKa = 3.52GTAVAGTGSTGTTSGVFRR243 pKa = 11.84WTADD247 pKa = 2.66TGMVDD252 pKa = 2.79IGTLGGIYY260 pKa = 9.93TSVAAMSGDD269 pKa = 3.47GNVLVGQSTTVGEE282 pKa = 4.47SDD284 pKa = 3.42YY285 pKa = 11.24HH286 pKa = 7.22AYY288 pKa = 10.22RR289 pKa = 11.84YY290 pKa = 8.91VASTSTMTDD299 pKa = 2.84LGTLGGTYY307 pKa = 9.98SYY309 pKa = 11.63ASDD312 pKa = 3.64LTADD316 pKa = 3.71GSIVVGRR323 pKa = 11.84ASDD326 pKa = 3.52SASAYY331 pKa = 10.43HH332 pKa = 6.04GFVWSEE338 pKa = 3.74ATGMQTVEE346 pKa = 4.31DD347 pKa = 4.18WLTAHH352 pKa = 7.4GATLEE357 pKa = 4.05GDD359 pKa = 3.61MTTSADD365 pKa = 3.99KK366 pKa = 10.72ISADD370 pKa = 3.07GSVIVGSTTSGSTYY384 pKa = 9.18IARR387 pKa = 11.84VVTTGSDD394 pKa = 3.36DD395 pKa = 3.61STGGDD400 pKa = 3.09SGGGDD405 pKa = 3.8SEE407 pKa = 5.29CNADD411 pKa = 3.47VCDD414 pKa = 4.27GGGEE418 pKa = 4.09SGIIDD423 pKa = 3.6TAKK426 pKa = 10.52YY427 pKa = 9.65FPTVASANNMIVQSGVNSADD447 pKa = 3.14TIMFGAQGAPMRR459 pKa = 11.84NLLKK463 pKa = 10.05TGQKK467 pKa = 10.0SVWGTVDD474 pKa = 3.41GGYY477 pKa = 10.55DD478 pKa = 4.11DD479 pKa = 5.82SDD481 pKa = 3.91HH482 pKa = 7.27ADD484 pKa = 3.36GGLALGDD491 pKa = 3.66FGFGYY496 pKa = 10.79GIAEE500 pKa = 4.16GVTARR505 pKa = 11.84FSAGMTYY512 pKa = 10.19TDD514 pKa = 3.33QDD516 pKa = 3.6LDD518 pKa = 3.46AGGDD522 pKa = 3.34VRR524 pKa = 11.84QRR526 pKa = 11.84GFYY529 pKa = 10.58LSPEE533 pKa = 3.99VSADD537 pKa = 3.29LGRR540 pKa = 11.84NVYY543 pKa = 7.73MTVGGYY549 pKa = 8.24WGRR552 pKa = 11.84SSIDD556 pKa = 2.95SRR558 pKa = 11.84RR559 pKa = 11.84GYY561 pKa = 11.27ASGAITDD568 pKa = 3.68YY569 pKa = 11.38SYY571 pKa = 12.1GDD573 pKa = 3.67TNAEE577 pKa = 3.61TWGAKK582 pKa = 9.1IRR584 pKa = 11.84FDD586 pKa = 3.61WLNAATIADD595 pKa = 4.1TAITPYY601 pKa = 10.89AGLSYY606 pKa = 11.16AHH608 pKa = 6.19TRR610 pKa = 11.84VDD612 pKa = 3.56GFSEE616 pKa = 4.14TGGAFPVEE624 pKa = 4.21FDD626 pKa = 3.74GSSDD630 pKa = 3.08HH631 pKa = 6.65ATIARR636 pKa = 11.84LGSDD640 pKa = 3.85FVRR643 pKa = 11.84PLNDD647 pKa = 3.37TVRR650 pKa = 11.84LLAKK654 pKa = 10.58AEE656 pKa = 3.94LDD658 pKa = 3.76YY659 pKa = 11.43QFEE662 pKa = 4.15NHH664 pKa = 6.24AAATTGTLTGISDD677 pKa = 4.48FDD679 pKa = 5.42LEE681 pKa = 4.54GQDD684 pKa = 4.99LQQFWVRR691 pKa = 11.84GGLGAEE697 pKa = 3.87FDD699 pKa = 3.79VGKK702 pKa = 10.65GVASFMVNATTKK714 pKa = 10.62GQDD717 pKa = 3.08PTVWLRR723 pKa = 11.84SNYY726 pKa = 7.13TVKK729 pKa = 10.76FF730 pKa = 3.7
Molecular weight: 75.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A386E6G1|A0A386E6G1_9RHIZ Amino acid ABC transporter substrate-binding protein OS=Neorhizobium sp. NCHU2750 OX=1825976 GN=NCHU2750_45240 PE=3 SV=1
MM1 pKa = 7.66PGKK4 pKa = 10.03SPSSGGVAVKK14 pKa = 10.01GVRR17 pKa = 11.84RR18 pKa = 11.84SALRR22 pKa = 11.84MASTLAAGACGALLAKK38 pKa = 10.38LVGVPLPFLLGPLVFGAAATLAGAPVLTVPYY69 pKa = 9.56GRR71 pKa = 11.84QIGQVVVGVSIGLRR85 pKa = 11.84FIPSVAVATLTLIPVMVAVTVLVIVATSIAALLLMRR121 pKa = 11.84LGGVDD126 pKa = 3.31RR127 pKa = 11.84EE128 pKa = 4.4TAFFATAAAGLAEE141 pKa = 4.33MAVVAQNKK149 pKa = 8.34GANPDD154 pKa = 3.08IVAVVHH160 pKa = 6.51LIRR163 pKa = 11.84VTSIVTSVPILLAIFGHH180 pKa = 6.96PGDD183 pKa = 4.3VPPVAVPLAAEE194 pKa = 4.62LLPLAGLLVVSGIAAYY210 pKa = 10.06LVAPFGMPNTWLLIPTLIGALVALSGFGPFAVPKK244 pKa = 10.45LLLNLAQIVIGTWIGCRR261 pKa = 11.84FRR263 pKa = 11.84RR264 pKa = 11.84EE265 pKa = 3.43IVARR269 pKa = 11.84LPRR272 pKa = 11.84VSLAAVATTAFLLASAALIASLVCWMTGLPFSTAFLSISPAGVTEE317 pKa = 4.21MVLTATAMHH326 pKa = 7.1LDD328 pKa = 3.5AATVTGFQIMRR339 pKa = 11.84IAVVMTTIPLVFRR352 pKa = 11.84LFAYY356 pKa = 10.62LSAPDD361 pKa = 4.45ADD363 pKa = 5.38RR364 pKa = 11.84IDD366 pKa = 3.6APRR369 pKa = 11.84PP370 pKa = 3.45
MM1 pKa = 7.66PGKK4 pKa = 10.03SPSSGGVAVKK14 pKa = 10.01GVRR17 pKa = 11.84RR18 pKa = 11.84SALRR22 pKa = 11.84MASTLAAGACGALLAKK38 pKa = 10.38LVGVPLPFLLGPLVFGAAATLAGAPVLTVPYY69 pKa = 9.56GRR71 pKa = 11.84QIGQVVVGVSIGLRR85 pKa = 11.84FIPSVAVATLTLIPVMVAVTVLVIVATSIAALLLMRR121 pKa = 11.84LGGVDD126 pKa = 3.31RR127 pKa = 11.84EE128 pKa = 4.4TAFFATAAAGLAEE141 pKa = 4.33MAVVAQNKK149 pKa = 8.34GANPDD154 pKa = 3.08IVAVVHH160 pKa = 6.51LIRR163 pKa = 11.84VTSIVTSVPILLAIFGHH180 pKa = 6.96PGDD183 pKa = 4.3VPPVAVPLAAEE194 pKa = 4.62LLPLAGLLVVSGIAAYY210 pKa = 10.06LVAPFGMPNTWLLIPTLIGALVALSGFGPFAVPKK244 pKa = 10.45LLLNLAQIVIGTWIGCRR261 pKa = 11.84FRR263 pKa = 11.84RR264 pKa = 11.84EE265 pKa = 3.43IVARR269 pKa = 11.84LPRR272 pKa = 11.84VSLAAVATTAFLLASAALIASLVCWMTGLPFSTAFLSISPAGVTEE317 pKa = 4.21MVLTATAMHH326 pKa = 7.1LDD328 pKa = 3.5AATVTGFQIMRR339 pKa = 11.84IAVVMTTIPLVFRR352 pKa = 11.84LFAYY356 pKa = 10.62LSAPDD361 pKa = 4.45ADD363 pKa = 5.38RR364 pKa = 11.84IDD366 pKa = 3.6APRR369 pKa = 11.84PP370 pKa = 3.45
Molecular weight: 37.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1837426 |
29 |
2828 |
311.4 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.821 ± 0.042 | 0.782 ± 0.01 |
5.788 ± 0.032 | 5.61 ± 0.024 |
3.872 ± 0.018 | 8.319 ± 0.031 |
1.974 ± 0.015 | 5.802 ± 0.024 |
3.83 ± 0.026 | 9.851 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.692 ± 0.016 | 2.864 ± 0.019 |
4.854 ± 0.022 | 3.183 ± 0.025 |
6.482 ± 0.037 | 6.016 ± 0.026 |
5.434 ± 0.028 | 7.25 ± 0.028 |
1.253 ± 0.013 | 2.322 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
