
Bat circovirus POA/2012/VI
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Bat associated cyclovirus 10
Average proteome isoelectric point is 9.17
Get precalculated fractions of proteins
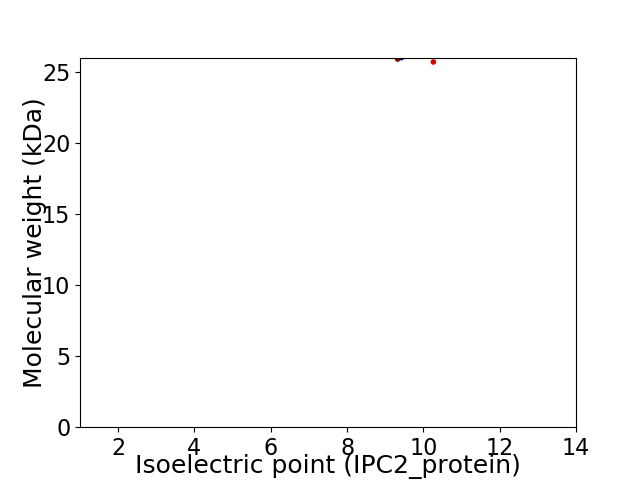
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
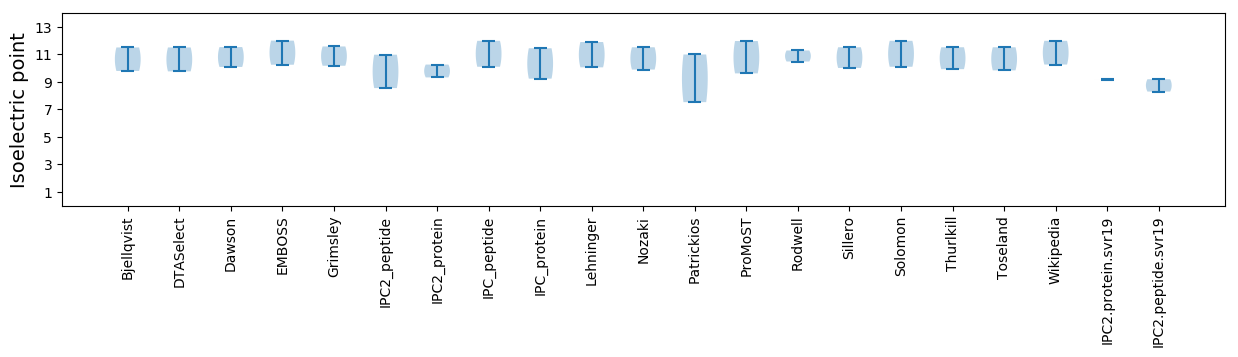
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0YUE0|A0A0A0YUE0_9CIRC Capsid OS=Bat circovirus POA/2012/VI OX=1572239 GN=Cap PE=4 SV=1
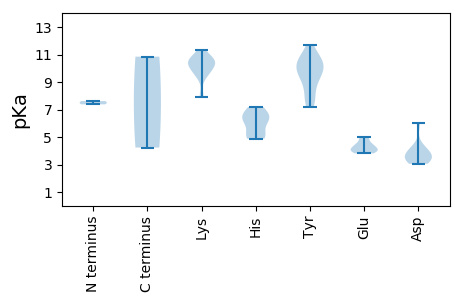
MM1 pKa = 7.62AFRR4 pKa = 11.84RR5 pKa = 11.84FRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84MPLRR15 pKa = 11.84RR16 pKa = 11.84NRR18 pKa = 11.84FRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84IFRR25 pKa = 11.84HH26 pKa = 4.88RR27 pKa = 11.84LRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84VVRR34 pKa = 11.84RR35 pKa = 11.84TPTLYY40 pKa = 10.89VKK42 pKa = 9.38LTRR45 pKa = 11.84TIQIAIPPSVEE56 pKa = 3.96FNQPLHH62 pKa = 6.64VSLNDD67 pKa = 3.48FAEE70 pKa = 5.04HH71 pKa = 6.88INLAPNFEE79 pKa = 4.3RR80 pKa = 11.84VKK82 pKa = 10.84ALRR85 pKa = 11.84QTVRR89 pKa = 11.84VYY91 pKa = 10.06PQQNVSNTSTSRR103 pKa = 11.84VGNYY107 pKa = 10.15CMLPYY112 pKa = 10.0HH113 pKa = 6.51KK114 pKa = 9.68PVPTSLINFPTALSVDD130 pKa = 3.25KK131 pKa = 11.18AKK133 pKa = 10.75VFRR136 pKa = 11.84CTQKK140 pKa = 10.55GRR142 pKa = 11.84MSFVPATRR150 pKa = 11.84LDD152 pKa = 3.59ADD154 pKa = 3.94GVGGNQTIRR163 pKa = 11.84TDD165 pKa = 3.05WRR167 pKa = 11.84PEE169 pKa = 3.81LEE171 pKa = 4.32IGSTATNVIMYY182 pKa = 8.82TGMIVFEE189 pKa = 4.92NINAGLTGTTAYY201 pKa = 8.76YY202 pKa = 9.95TVVQDD207 pKa = 4.33LYY209 pKa = 11.69VRR211 pKa = 11.84YY212 pKa = 9.71KK213 pKa = 9.84NQRR216 pKa = 11.84SFII219 pKa = 4.23
MM1 pKa = 7.62AFRR4 pKa = 11.84RR5 pKa = 11.84FRR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84MPLRR15 pKa = 11.84RR16 pKa = 11.84NRR18 pKa = 11.84FRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84IFRR25 pKa = 11.84HH26 pKa = 4.88RR27 pKa = 11.84LRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84VVRR34 pKa = 11.84RR35 pKa = 11.84TPTLYY40 pKa = 10.89VKK42 pKa = 9.38LTRR45 pKa = 11.84TIQIAIPPSVEE56 pKa = 3.96FNQPLHH62 pKa = 6.64VSLNDD67 pKa = 3.48FAEE70 pKa = 5.04HH71 pKa = 6.88INLAPNFEE79 pKa = 4.3RR80 pKa = 11.84VKK82 pKa = 10.84ALRR85 pKa = 11.84QTVRR89 pKa = 11.84VYY91 pKa = 10.06PQQNVSNTSTSRR103 pKa = 11.84VGNYY107 pKa = 10.15CMLPYY112 pKa = 10.0HH113 pKa = 6.51KK114 pKa = 9.68PVPTSLINFPTALSVDD130 pKa = 3.25KK131 pKa = 11.18AKK133 pKa = 10.75VFRR136 pKa = 11.84CTQKK140 pKa = 10.55GRR142 pKa = 11.84MSFVPATRR150 pKa = 11.84LDD152 pKa = 3.59ADD154 pKa = 3.94GVGGNQTIRR163 pKa = 11.84TDD165 pKa = 3.05WRR167 pKa = 11.84PEE169 pKa = 3.81LEE171 pKa = 4.32IGSTATNVIMYY182 pKa = 8.82TGMIVFEE189 pKa = 4.92NINAGLTGTTAYY201 pKa = 8.76YY202 pKa = 9.95TVVQDD207 pKa = 4.33LYY209 pKa = 11.69VRR211 pKa = 11.84YY212 pKa = 9.71KK213 pKa = 9.84NQRR216 pKa = 11.84SFII219 pKa = 4.23
Molecular weight: 25.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0YUE0|A0A0A0YUE0_9CIRC Capsid OS=Bat circovirus POA/2012/VI OX=1572239 GN=Cap PE=4 SV=1
MM1 pKa = 7.38SNSTVRR7 pKa = 11.84RR8 pKa = 11.84FCFTWNNYY16 pKa = 7.15TVADD20 pKa = 4.1TLTVKK25 pKa = 10.69DD26 pKa = 6.02FFIQHH31 pKa = 6.26CKK33 pKa = 10.2YY34 pKa = 10.51GIAGEE39 pKa = 4.53EE40 pKa = 3.96IAPTTGTPHH49 pKa = 6.23LQGFANLSKK58 pKa = 10.6PMRR61 pKa = 11.84FSTIKK66 pKa = 10.06KK67 pKa = 8.07HH68 pKa = 5.0LHH70 pKa = 5.39NGIHH74 pKa = 6.11IEE76 pKa = 4.0KK77 pKa = 10.66ANGSDD82 pKa = 3.6EE83 pKa = 4.48QNQAYY88 pKa = 9.27CKK90 pKa = 10.51KK91 pKa = 9.91SGTWFEE97 pKa = 4.06QGTPCSQGSRR107 pKa = 11.84SDD109 pKa = 3.53LADD112 pKa = 3.38VVSAIQNGTNTTQAIAQLYY131 pKa = 7.72PIAYY135 pKa = 7.69IKK137 pKa = 10.26YY138 pKa = 7.88YY139 pKa = 10.66KK140 pKa = 10.33GINEE144 pKa = 4.05YY145 pKa = 11.01LKK147 pKa = 9.42IQHH150 pKa = 7.19PITPRR155 pKa = 11.84RR156 pKa = 11.84HH157 pKa = 5.04KK158 pKa = 9.46TWVYY162 pKa = 9.69YY163 pKa = 9.85YY164 pKa = 9.15WGPPGTGKK172 pKa = 10.03SRR174 pKa = 11.84RR175 pKa = 11.84ALKK178 pKa = 10.01KK179 pKa = 10.4RR180 pKa = 11.84RR181 pKa = 11.84VSTQKK186 pKa = 10.48VSTTSQGACGGTDD199 pKa = 3.19TSNKK203 pKa = 7.89RR204 pKa = 11.84TSSSTTSTGGSNTTNYY220 pKa = 10.79SKK222 pKa = 11.34LLTDD226 pKa = 4.0TLTKK230 pKa = 10.8YY231 pKa = 10.43KK232 pKa = 10.85
MM1 pKa = 7.38SNSTVRR7 pKa = 11.84RR8 pKa = 11.84FCFTWNNYY16 pKa = 7.15TVADD20 pKa = 4.1TLTVKK25 pKa = 10.69DD26 pKa = 6.02FFIQHH31 pKa = 6.26CKK33 pKa = 10.2YY34 pKa = 10.51GIAGEE39 pKa = 4.53EE40 pKa = 3.96IAPTTGTPHH49 pKa = 6.23LQGFANLSKK58 pKa = 10.6PMRR61 pKa = 11.84FSTIKK66 pKa = 10.06KK67 pKa = 8.07HH68 pKa = 5.0LHH70 pKa = 5.39NGIHH74 pKa = 6.11IEE76 pKa = 4.0KK77 pKa = 10.66ANGSDD82 pKa = 3.6EE83 pKa = 4.48QNQAYY88 pKa = 9.27CKK90 pKa = 10.51KK91 pKa = 9.91SGTWFEE97 pKa = 4.06QGTPCSQGSRR107 pKa = 11.84SDD109 pKa = 3.53LADD112 pKa = 3.38VVSAIQNGTNTTQAIAQLYY131 pKa = 7.72PIAYY135 pKa = 7.69IKK137 pKa = 10.26YY138 pKa = 7.88YY139 pKa = 10.66KK140 pKa = 10.33GINEE144 pKa = 4.05YY145 pKa = 11.01LKK147 pKa = 9.42IQHH150 pKa = 7.19PITPRR155 pKa = 11.84RR156 pKa = 11.84HH157 pKa = 5.04KK158 pKa = 9.46TWVYY162 pKa = 9.69YY163 pKa = 9.85YY164 pKa = 9.15WGPPGTGKK172 pKa = 10.03SRR174 pKa = 11.84RR175 pKa = 11.84ALKK178 pKa = 10.01KK179 pKa = 10.4RR180 pKa = 11.84RR181 pKa = 11.84VSTQKK186 pKa = 10.48VSTTSQGACGGTDD199 pKa = 3.19TSNKK203 pKa = 7.89RR204 pKa = 11.84TSSSTTSTGGSNTTNYY220 pKa = 10.79SKK222 pKa = 11.34LLTDD226 pKa = 4.0TLTKK230 pKa = 10.8YY231 pKa = 10.43KK232 pKa = 10.85
Molecular weight: 25.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
451 |
219 |
232 |
225.5 |
25.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.543 ± 0.043 | 1.552 ± 0.435 |
2.882 ± 0.097 | 2.661 ± 0.054 |
4.213 ± 0.862 | 6.208 ± 1.429 |
2.439 ± 0.417 | 5.543 ± 0.043 |
5.987 ± 1.899 | 5.765 ± 0.738 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.774 ± 0.657 | 5.987 ± 0.276 |
4.878 ± 0.72 | 4.656 ± 0.372 |
9.313 ± 3.296 | 6.874 ± 1.571 |
11.308 ± 1.481 | 6.208 ± 1.99 |
1.33 ± 0.284 | 4.878 ± 0.523 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
