
Dysgonomonas alginatilytica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
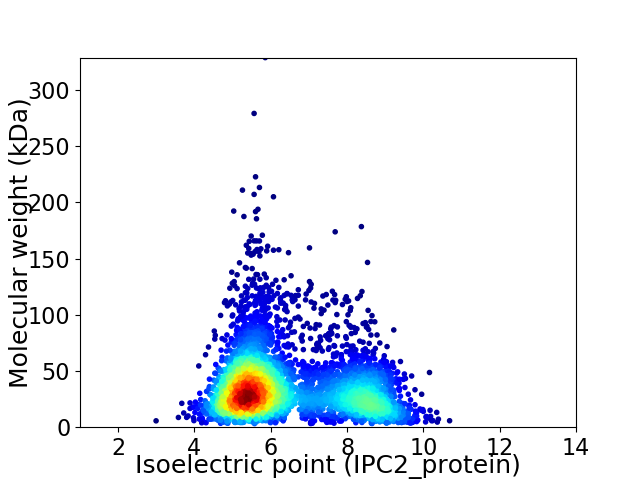
Virtual 2D-PAGE plot for 4322 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V3PRK5|A0A2V3PRK5_9BACT Uncharacterized protein OS=Dysgonomonas alginatilytica OX=1605892 GN=CLV62_10813 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.21ISPNKK7 pKa = 9.62FVSLSYY13 pKa = 10.8DD14 pKa = 3.62LNVGEE19 pKa = 4.67GDD21 pKa = 3.42EE22 pKa = 4.38KK23 pKa = 11.42EE24 pKa = 4.5LMEE27 pKa = 4.92KK28 pKa = 9.17ATPEE32 pKa = 3.81QPLEE36 pKa = 4.37FIFGTNSMLEE46 pKa = 4.03AFEE49 pKa = 4.87KK50 pKa = 11.02NVDD53 pKa = 3.51GLAEE57 pKa = 4.37GDD59 pKa = 3.78SFDD62 pKa = 6.45FILTPDD68 pKa = 3.47QAYY71 pKa = 10.41GEE73 pKa = 4.48YY74 pKa = 10.26DD75 pKa = 3.75DD76 pKa = 5.02EE77 pKa = 5.52HH78 pKa = 8.85LVDD81 pKa = 4.02LPKK84 pKa = 11.0NIFEE88 pKa = 4.37VDD90 pKa = 3.53GKK92 pKa = 10.87VDD94 pKa = 3.45EE95 pKa = 4.63SVVFEE100 pKa = 4.66GNTIPMMDD108 pKa = 3.38SNGNRR113 pKa = 11.84LNGSVVSVSDD123 pKa = 3.95DD124 pKa = 3.39TVQMDD129 pKa = 4.73FNHH132 pKa = 6.85PLAGEE137 pKa = 4.2TLHH140 pKa = 6.11FTGNVLSIRR149 pKa = 11.84EE150 pKa = 4.01ASAEE154 pKa = 3.94EE155 pKa = 3.93IAALFAPQGGGCGSGCGCGSSDD177 pKa = 3.55EE178 pKa = 4.89SEE180 pKa = 4.62GCGSGCGCGSSDD192 pKa = 3.38EE193 pKa = 4.92SEE195 pKa = 4.28GSSCGSGCGCHH206 pKa = 6.4
MM1 pKa = 7.63KK2 pKa = 10.21ISPNKK7 pKa = 9.62FVSLSYY13 pKa = 10.8DD14 pKa = 3.62LNVGEE19 pKa = 4.67GDD21 pKa = 3.42EE22 pKa = 4.38KK23 pKa = 11.42EE24 pKa = 4.5LMEE27 pKa = 4.92KK28 pKa = 9.17ATPEE32 pKa = 3.81QPLEE36 pKa = 4.37FIFGTNSMLEE46 pKa = 4.03AFEE49 pKa = 4.87KK50 pKa = 11.02NVDD53 pKa = 3.51GLAEE57 pKa = 4.37GDD59 pKa = 3.78SFDD62 pKa = 6.45FILTPDD68 pKa = 3.47QAYY71 pKa = 10.41GEE73 pKa = 4.48YY74 pKa = 10.26DD75 pKa = 3.75DD76 pKa = 5.02EE77 pKa = 5.52HH78 pKa = 8.85LVDD81 pKa = 4.02LPKK84 pKa = 11.0NIFEE88 pKa = 4.37VDD90 pKa = 3.53GKK92 pKa = 10.87VDD94 pKa = 3.45EE95 pKa = 4.63SVVFEE100 pKa = 4.66GNTIPMMDD108 pKa = 3.38SNGNRR113 pKa = 11.84LNGSVVSVSDD123 pKa = 3.95DD124 pKa = 3.39TVQMDD129 pKa = 4.73FNHH132 pKa = 6.85PLAGEE137 pKa = 4.2TLHH140 pKa = 6.11FTGNVLSIRR149 pKa = 11.84EE150 pKa = 4.01ASAEE154 pKa = 3.94EE155 pKa = 3.93IAALFAPQGGGCGSGCGCGSSDD177 pKa = 3.55EE178 pKa = 4.89SEE180 pKa = 4.62GCGSGCGCGSSDD192 pKa = 3.38EE193 pKa = 4.92SEE195 pKa = 4.28GSSCGSGCGCHH206 pKa = 6.4
Molecular weight: 21.63 kDa
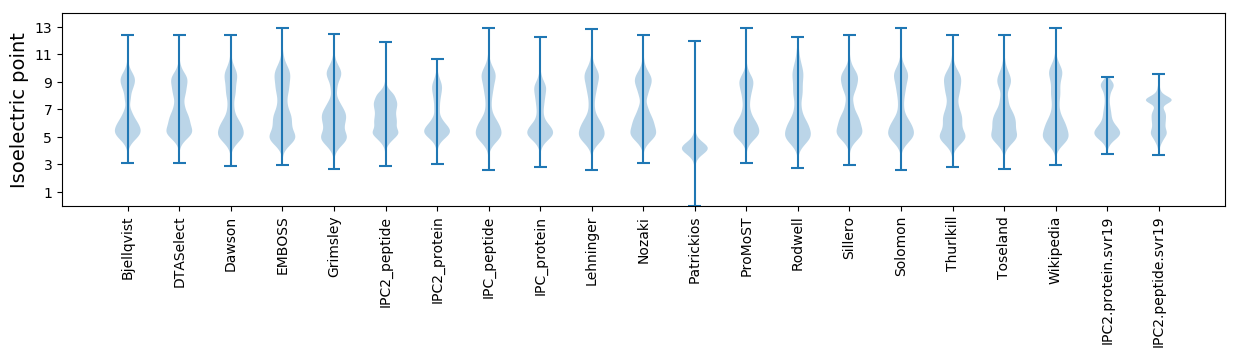
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V3PJX7|A0A2V3PJX7_9BACT Uncharacterized protein OS=Dysgonomonas alginatilytica OX=1605892 GN=CLV62_15220 PE=4 SV=1
MM1 pKa = 7.29ATLTFYY7 pKa = 11.1LRR9 pKa = 11.84ASSRR13 pKa = 11.84IGCHH17 pKa = 6.53AGTLCFRR24 pKa = 11.84IIHH27 pKa = 5.83ARR29 pKa = 11.84RR30 pKa = 11.84VLSHH34 pKa = 5.47STNIRR39 pKa = 11.84LYY41 pKa = 10.03CRR43 pKa = 11.84EE44 pKa = 3.79WDD46 pKa = 3.57STSRR50 pKa = 11.84RR51 pKa = 11.84IVLSDD56 pKa = 3.18SSEE59 pKa = 3.81EE60 pKa = 4.06RR61 pKa = 11.84ITYY64 pKa = 9.87LNKK67 pKa = 10.38AEE69 pKa = 3.98QQQQNIEE76 pKa = 3.8QSFAHH81 pKa = 7.1IIHH84 pKa = 6.31NLEE87 pKa = 3.33EE88 pKa = 4.11SGRR91 pKa = 11.84YY92 pKa = 8.58RR93 pKa = 11.84VEE95 pKa = 6.13DD96 pKa = 2.85IMGRR100 pKa = 11.84YY101 pKa = 9.11RR102 pKa = 11.84LRR104 pKa = 11.84LDD106 pKa = 3.51SHH108 pKa = 6.21WLYY111 pKa = 10.99HH112 pKa = 5.1FTGQQSHH119 pKa = 5.68TLIASGRR126 pKa = 11.84EE127 pKa = 3.7RR128 pKa = 11.84TARR131 pKa = 11.84AYY133 pKa = 8.14RR134 pKa = 11.84TAARR138 pKa = 11.84GLIAFNAGRR147 pKa = 11.84DD148 pKa = 3.8LPLKK152 pKa = 10.16HH153 pKa = 6.73LNSCLIKK160 pKa = 10.89DD161 pKa = 4.49FEE163 pKa = 5.23NDD165 pKa = 3.24MKK167 pKa = 11.27HH168 pKa = 6.6KK169 pKa = 10.69GKK171 pKa = 8.13TLNTISFYY179 pKa = 10.26MRR181 pKa = 11.84NLRR184 pKa = 11.84AIYY187 pKa = 10.18NKK189 pKa = 10.44AIACKK194 pKa = 10.09RR195 pKa = 11.84IADD198 pKa = 3.78TGQNPFSDD206 pKa = 4.32VYY208 pKa = 10.99TGVQKK213 pKa = 9.54TRR215 pKa = 11.84KK216 pKa = 9.28RR217 pKa = 11.84SLTMDD222 pKa = 3.26QLKK225 pKa = 8.51TLNDD229 pKa = 3.86LDD231 pKa = 4.67LSLGSGTTAGAKK243 pKa = 9.53EE244 pKa = 4.17GSSVRR249 pKa = 11.84DD250 pKa = 3.28QEE252 pKa = 4.13IFRR255 pKa = 11.84GLDD258 pKa = 2.85AARR261 pKa = 11.84RR262 pKa = 11.84LFLFCFHH269 pKa = 6.83ARR271 pKa = 11.84GMSFVDD277 pKa = 3.74LAYY280 pKa = 10.55LRR282 pKa = 11.84KK283 pKa = 10.12EE284 pKa = 4.04NIQGKK289 pKa = 9.05RR290 pKa = 11.84IRR292 pKa = 11.84YY293 pKa = 7.09YY294 pKa = 10.19RR295 pKa = 11.84KK296 pKa = 8.28KK297 pKa = 8.92TGGLIEE303 pKa = 5.01LKK305 pKa = 8.82ITPVMQEE312 pKa = 4.19IIHH315 pKa = 5.88SFSLQTRR322 pKa = 11.84DD323 pKa = 2.97SLYY326 pKa = 10.04IFPILVSPHH335 pKa = 6.02KK336 pKa = 9.81PLRR339 pKa = 11.84LQYY342 pKa = 10.73EE343 pKa = 4.19SGLRR347 pKa = 11.84LQNNRR352 pKa = 11.84LKK354 pKa = 10.78RR355 pKa = 11.84LARR358 pKa = 11.84LAGIHH363 pKa = 5.97SAVSTHH369 pKa = 5.53VSRR372 pKa = 11.84HH373 pKa = 4.32SWASAAKK380 pKa = 10.07KK381 pKa = 10.14EE382 pKa = 4.13NLPVIPVRR390 pKa = 11.84SMPRR394 pKa = 11.84RR395 pKa = 3.52
MM1 pKa = 7.29ATLTFYY7 pKa = 11.1LRR9 pKa = 11.84ASSRR13 pKa = 11.84IGCHH17 pKa = 6.53AGTLCFRR24 pKa = 11.84IIHH27 pKa = 5.83ARR29 pKa = 11.84RR30 pKa = 11.84VLSHH34 pKa = 5.47STNIRR39 pKa = 11.84LYY41 pKa = 10.03CRR43 pKa = 11.84EE44 pKa = 3.79WDD46 pKa = 3.57STSRR50 pKa = 11.84RR51 pKa = 11.84IVLSDD56 pKa = 3.18SSEE59 pKa = 3.81EE60 pKa = 4.06RR61 pKa = 11.84ITYY64 pKa = 9.87LNKK67 pKa = 10.38AEE69 pKa = 3.98QQQQNIEE76 pKa = 3.8QSFAHH81 pKa = 7.1IIHH84 pKa = 6.31NLEE87 pKa = 3.33EE88 pKa = 4.11SGRR91 pKa = 11.84YY92 pKa = 8.58RR93 pKa = 11.84VEE95 pKa = 6.13DD96 pKa = 2.85IMGRR100 pKa = 11.84YY101 pKa = 9.11RR102 pKa = 11.84LRR104 pKa = 11.84LDD106 pKa = 3.51SHH108 pKa = 6.21WLYY111 pKa = 10.99HH112 pKa = 5.1FTGQQSHH119 pKa = 5.68TLIASGRR126 pKa = 11.84EE127 pKa = 3.7RR128 pKa = 11.84TARR131 pKa = 11.84AYY133 pKa = 8.14RR134 pKa = 11.84TAARR138 pKa = 11.84GLIAFNAGRR147 pKa = 11.84DD148 pKa = 3.8LPLKK152 pKa = 10.16HH153 pKa = 6.73LNSCLIKK160 pKa = 10.89DD161 pKa = 4.49FEE163 pKa = 5.23NDD165 pKa = 3.24MKK167 pKa = 11.27HH168 pKa = 6.6KK169 pKa = 10.69GKK171 pKa = 8.13TLNTISFYY179 pKa = 10.26MRR181 pKa = 11.84NLRR184 pKa = 11.84AIYY187 pKa = 10.18NKK189 pKa = 10.44AIACKK194 pKa = 10.09RR195 pKa = 11.84IADD198 pKa = 3.78TGQNPFSDD206 pKa = 4.32VYY208 pKa = 10.99TGVQKK213 pKa = 9.54TRR215 pKa = 11.84KK216 pKa = 9.28RR217 pKa = 11.84SLTMDD222 pKa = 3.26QLKK225 pKa = 8.51TLNDD229 pKa = 3.86LDD231 pKa = 4.67LSLGSGTTAGAKK243 pKa = 9.53EE244 pKa = 4.17GSSVRR249 pKa = 11.84DD250 pKa = 3.28QEE252 pKa = 4.13IFRR255 pKa = 11.84GLDD258 pKa = 2.85AARR261 pKa = 11.84RR262 pKa = 11.84LFLFCFHH269 pKa = 6.83ARR271 pKa = 11.84GMSFVDD277 pKa = 3.74LAYY280 pKa = 10.55LRR282 pKa = 11.84KK283 pKa = 10.12EE284 pKa = 4.04NIQGKK289 pKa = 9.05RR290 pKa = 11.84IRR292 pKa = 11.84YY293 pKa = 7.09YY294 pKa = 10.19RR295 pKa = 11.84KK296 pKa = 8.28KK297 pKa = 8.92TGGLIEE303 pKa = 5.01LKK305 pKa = 8.82ITPVMQEE312 pKa = 4.19IIHH315 pKa = 5.88SFSLQTRR322 pKa = 11.84DD323 pKa = 2.97SLYY326 pKa = 10.04IFPILVSPHH335 pKa = 6.02KK336 pKa = 9.81PLRR339 pKa = 11.84LQYY342 pKa = 10.73EE343 pKa = 4.19SGLRR347 pKa = 11.84LQNNRR352 pKa = 11.84LKK354 pKa = 10.78RR355 pKa = 11.84LARR358 pKa = 11.84LAGIHH363 pKa = 5.97SAVSTHH369 pKa = 5.53VSRR372 pKa = 11.84HH373 pKa = 4.32SWASAAKK380 pKa = 10.07KK381 pKa = 10.14EE382 pKa = 4.13NLPVIPVRR390 pKa = 11.84SMPRR394 pKa = 11.84RR395 pKa = 3.52
Molecular weight: 45.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1489738 |
29 |
2897 |
344.7 |
38.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.255 ± 0.036 | 0.955 ± 0.012 |
5.763 ± 0.023 | 6.211 ± 0.031 |
4.816 ± 0.029 | 6.421 ± 0.035 |
1.627 ± 0.015 | 8.071 ± 0.036 |
7.416 ± 0.03 | 9.055 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.016 | 5.972 ± 0.033 |
3.48 ± 0.02 | 3.38 ± 0.022 |
3.86 ± 0.024 | 6.813 ± 0.029 |
5.736 ± 0.028 | 6.007 ± 0.029 |
1.219 ± 0.015 | 4.596 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
