
Runella sp. SP2
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Runella; unclassified Runella
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
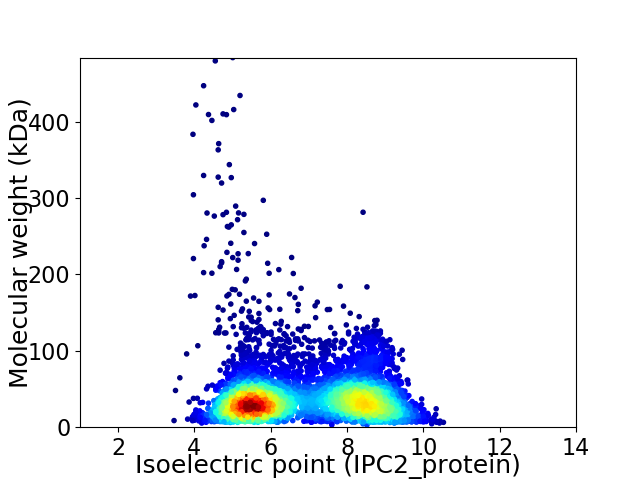
Virtual 2D-PAGE plot for 5998 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G3GJR2|A0A3G3GJR2_9BACT FecR family protein OS=Runella sp. SP2 OX=2268026 GN=DTQ70_05275 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.37ALQTFIFVLSLFCIVKK18 pKa = 10.61GNAQDD23 pKa = 3.41VGINILNQPASVSQGSTNGRR43 pKa = 11.84IIIDD47 pKa = 3.17ICNFDD52 pKa = 4.22GGTTSTPLNRR62 pKa = 11.84IQPLITLPSALVDD75 pKa = 3.57TFVTAISFDD84 pKa = 3.05GWSIVSKK91 pKa = 10.53SGSTIRR97 pKa = 11.84FQNTVSITPGEE108 pKa = 4.31CKK110 pKa = 10.4QIVLGYY116 pKa = 10.25KK117 pKa = 9.51GVNVGGPLTVTGTIQFSGPQTSGNNPANDD146 pKa = 3.38NSTTSIAVTSAPVDD160 pKa = 3.49TDD162 pKa = 3.23GDD164 pKa = 4.53GIPDD168 pKa = 3.83STDD171 pKa = 3.7LDD173 pKa = 4.3DD174 pKa = 6.68DD175 pKa = 4.23NDD177 pKa = 5.09GILDD181 pKa = 3.91TVEE184 pKa = 3.91NAAVCSTVLFNGVANTDD201 pKa = 3.69CDD203 pKa = 4.01GDD205 pKa = 4.4GIPNYY210 pKa = 10.62LDD212 pKa = 4.76LDD214 pKa = 3.98SDD216 pKa = 3.89NDD218 pKa = 4.58GINDD222 pKa = 3.36VNEE225 pKa = 4.07AAGVDD230 pKa = 3.81VNGDD234 pKa = 3.47GFADD238 pKa = 4.28GTPDD242 pKa = 5.05AITGIPASAGNGLTPPNTDD261 pKa = 2.64GTGGTDD267 pKa = 4.17PYY269 pKa = 11.65DD270 pKa = 4.27LDD272 pKa = 4.39SDD274 pKa = 4.33NDD276 pKa = 4.14GQSDD280 pKa = 3.87LAEE283 pKa = 4.9GGLNASILDD292 pKa = 3.89PDD294 pKa = 3.91GDD296 pKa = 4.31GIVNCTANCDD306 pKa = 3.8PDD308 pKa = 4.02KK309 pKa = 11.39DD310 pKa = 4.52GILAPVDD317 pKa = 4.05GLPAAYY323 pKa = 10.1GDD325 pKa = 5.05AITNRR330 pKa = 11.84PPVANNDD337 pKa = 3.66TYY339 pKa = 11.58SILQGVILTDD349 pKa = 3.42NVLTNDD355 pKa = 3.9TDD357 pKa = 4.14PDD359 pKa = 4.14NNTLTVATTPTVAPTQGTVVLQTNGAFTYY388 pKa = 9.35TPTSATFTGQDD399 pKa = 3.35SFCYY403 pKa = 9.71LVCDD407 pKa = 3.52NGTPSLCKK415 pKa = 9.18TACATINIGAGIVDD429 pKa = 4.05LSISKK434 pKa = 10.29VLVGSKK440 pKa = 9.65IRR442 pKa = 11.84SLNDD446 pKa = 3.28TITYY450 pKa = 9.9RR451 pKa = 11.84IVVKK455 pKa = 9.79NTGNITATNIVVEE468 pKa = 4.68DD469 pKa = 3.73STTTGLQIISGTPTSGSFSGTNWTIPSLASGDD501 pKa = 3.75STTLVIQAKK510 pKa = 10.07VIAEE514 pKa = 3.79GVNFNFALIKK524 pKa = 10.85SADD527 pKa = 3.74QPDD530 pKa = 4.13TVATNDD536 pKa = 3.66TANDD540 pKa = 4.68CITVPIGLCSGQKK553 pKa = 10.0VEE555 pKa = 4.2VSVPSTYY562 pKa = 11.12TNVIWFKK569 pKa = 11.04NGAQIASGNVVLFSEE584 pKa = 4.54EE585 pKa = 3.53GTYY588 pKa = 10.54TFTASNVTCPVSGCCPLIIQSGTNCCPVQLCIPVTVVRR626 pKa = 11.84RR627 pKa = 11.84KK628 pKa = 9.75KK629 pKa = 10.53
MM1 pKa = 7.63KK2 pKa = 10.37ALQTFIFVLSLFCIVKK18 pKa = 10.61GNAQDD23 pKa = 3.41VGINILNQPASVSQGSTNGRR43 pKa = 11.84IIIDD47 pKa = 3.17ICNFDD52 pKa = 4.22GGTTSTPLNRR62 pKa = 11.84IQPLITLPSALVDD75 pKa = 3.57TFVTAISFDD84 pKa = 3.05GWSIVSKK91 pKa = 10.53SGSTIRR97 pKa = 11.84FQNTVSITPGEE108 pKa = 4.31CKK110 pKa = 10.4QIVLGYY116 pKa = 10.25KK117 pKa = 9.51GVNVGGPLTVTGTIQFSGPQTSGNNPANDD146 pKa = 3.38NSTTSIAVTSAPVDD160 pKa = 3.49TDD162 pKa = 3.23GDD164 pKa = 4.53GIPDD168 pKa = 3.83STDD171 pKa = 3.7LDD173 pKa = 4.3DD174 pKa = 6.68DD175 pKa = 4.23NDD177 pKa = 5.09GILDD181 pKa = 3.91TVEE184 pKa = 3.91NAAVCSTVLFNGVANTDD201 pKa = 3.69CDD203 pKa = 4.01GDD205 pKa = 4.4GIPNYY210 pKa = 10.62LDD212 pKa = 4.76LDD214 pKa = 3.98SDD216 pKa = 3.89NDD218 pKa = 4.58GINDD222 pKa = 3.36VNEE225 pKa = 4.07AAGVDD230 pKa = 3.81VNGDD234 pKa = 3.47GFADD238 pKa = 4.28GTPDD242 pKa = 5.05AITGIPASAGNGLTPPNTDD261 pKa = 2.64GTGGTDD267 pKa = 4.17PYY269 pKa = 11.65DD270 pKa = 4.27LDD272 pKa = 4.39SDD274 pKa = 4.33NDD276 pKa = 4.14GQSDD280 pKa = 3.87LAEE283 pKa = 4.9GGLNASILDD292 pKa = 3.89PDD294 pKa = 3.91GDD296 pKa = 4.31GIVNCTANCDD306 pKa = 3.8PDD308 pKa = 4.02KK309 pKa = 11.39DD310 pKa = 4.52GILAPVDD317 pKa = 4.05GLPAAYY323 pKa = 10.1GDD325 pKa = 5.05AITNRR330 pKa = 11.84PPVANNDD337 pKa = 3.66TYY339 pKa = 11.58SILQGVILTDD349 pKa = 3.42NVLTNDD355 pKa = 3.9TDD357 pKa = 4.14PDD359 pKa = 4.14NNTLTVATTPTVAPTQGTVVLQTNGAFTYY388 pKa = 9.35TPTSATFTGQDD399 pKa = 3.35SFCYY403 pKa = 9.71LVCDD407 pKa = 3.52NGTPSLCKK415 pKa = 9.18TACATINIGAGIVDD429 pKa = 4.05LSISKK434 pKa = 10.29VLVGSKK440 pKa = 9.65IRR442 pKa = 11.84SLNDD446 pKa = 3.28TITYY450 pKa = 9.9RR451 pKa = 11.84IVVKK455 pKa = 9.79NTGNITATNIVVEE468 pKa = 4.68DD469 pKa = 3.73STTTGLQIISGTPTSGSFSGTNWTIPSLASGDD501 pKa = 3.75STTLVIQAKK510 pKa = 10.07VIAEE514 pKa = 3.79GVNFNFALIKK524 pKa = 10.85SADD527 pKa = 3.74QPDD530 pKa = 4.13TVATNDD536 pKa = 3.66TANDD540 pKa = 4.68CITVPIGLCSGQKK553 pKa = 10.0VEE555 pKa = 4.2VSVPSTYY562 pKa = 11.12TNVIWFKK569 pKa = 11.04NGAQIASGNVVLFSEE584 pKa = 4.54EE585 pKa = 3.53GTYY588 pKa = 10.54TFTASNVTCPVSGCCPLIIQSGTNCCPVQLCIPVTVVRR626 pKa = 11.84RR627 pKa = 11.84KK628 pKa = 9.75KK629 pKa = 10.53
Molecular weight: 64.69 kDa
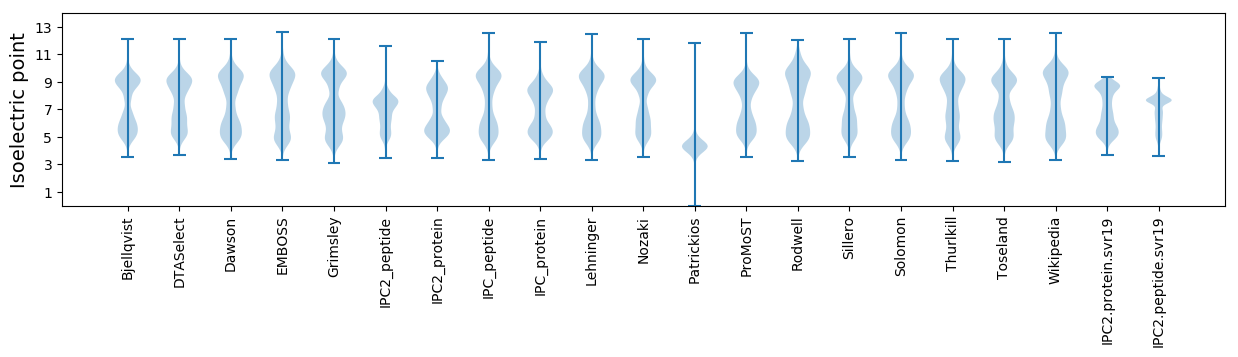
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G3GRY5|A0A3G3GRY5_9BACT Uncharacterized protein OS=Runella sp. SP2 OX=2268026 GN=DTQ70_18960 PE=4 SV=1
MM1 pKa = 7.26KK2 pKa = 10.3KK3 pKa = 9.77IVEE6 pKa = 4.15FLQLGEE12 pKa = 4.04VFGYY16 pKa = 8.46FLRR19 pKa = 11.84VFQKK23 pKa = 10.17PDD25 pKa = 3.38PSRR28 pKa = 11.84PSTTSLRR35 pKa = 11.84VMHH38 pKa = 7.16GINRR42 pKa = 11.84ISIVMFLFCLGVMLYY57 pKa = 10.62RR58 pKa = 11.84FFTRR62 pKa = 4.43
MM1 pKa = 7.26KK2 pKa = 10.3KK3 pKa = 9.77IVEE6 pKa = 4.15FLQLGEE12 pKa = 4.04VFGYY16 pKa = 8.46FLRR19 pKa = 11.84VFQKK23 pKa = 10.17PDD25 pKa = 3.38PSRR28 pKa = 11.84PSTTSLRR35 pKa = 11.84VMHH38 pKa = 7.16GINRR42 pKa = 11.84ISIVMFLFCLGVMLYY57 pKa = 10.62RR58 pKa = 11.84FFTRR62 pKa = 4.43
Molecular weight: 7.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2298142 |
25 |
4859 |
383.2 |
42.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.595 ± 0.033 | 0.9 ± 0.017 |
4.91 ± 0.025 | 5.628 ± 0.041 |
4.896 ± 0.027 | 7.102 ± 0.047 |
1.76 ± 0.02 | 6.175 ± 0.028 |
6.226 ± 0.043 | 9.506 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.02 | 5.307 ± 0.04 |
4.177 ± 0.02 | 4.103 ± 0.023 |
4.267 ± 0.03 | 6.562 ± 0.043 |
6.627 ± 0.069 | 6.886 ± 0.034 |
1.379 ± 0.013 | 3.899 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
