
Roseovarius pacificus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseovarius
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
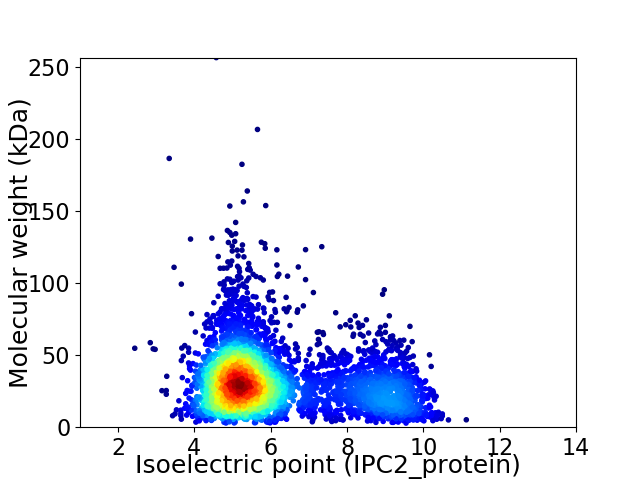
Virtual 2D-PAGE plot for 4424 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6WL23|A0A1M6WL23_9RHOB LuxR family transcriptional regulator OS=Roseovarius pacificus OX=337701 GN=SAMN05444398_10162 PE=4 SV=1
MM1 pKa = 7.6TIADD5 pKa = 4.36TMTLDD10 pKa = 3.57ALAQIAHH17 pKa = 5.05QTYY20 pKa = 6.96QQRR23 pKa = 11.84YY24 pKa = 8.17NPDD27 pKa = 2.69GSINTGSLNDD37 pKa = 4.04PVAGSNGSFYY47 pKa = 10.93TEE49 pKa = 3.28ITQIVNGPLGFQARR63 pKa = 11.84AFFNEE68 pKa = 4.09TTNEE72 pKa = 3.73LVIAFTGTEE81 pKa = 4.23GFPNDD86 pKa = 3.86PAEE89 pKa = 4.14FLPDD93 pKa = 3.95FLADD97 pKa = 4.33FSLAVAGVSPQDD109 pKa = 3.51LAAQAFIKK117 pKa = 10.52QATTLAQLEE126 pKa = 4.35AGPFGSFDD134 pKa = 3.35VTYY137 pKa = 10.39VGHH140 pKa = 6.31SLGGFLAQTASASGPEE156 pKa = 4.17GEE158 pKa = 4.17VVVFNAPGAGGFLGLPEE175 pKa = 4.13NHH177 pKa = 7.32PFPEE181 pKa = 4.55DD182 pKa = 3.09NFTYY186 pKa = 10.44VYY188 pKa = 10.67SDD190 pKa = 3.55PSDD193 pKa = 3.06WGAMGGAIHH202 pKa = 6.73SAGRR206 pKa = 11.84PLSDD210 pKa = 3.57NIYY213 pKa = 9.0YY214 pKa = 10.3VPGAEE219 pKa = 4.17GHH221 pKa = 7.15DD222 pKa = 3.11ISTPDD227 pKa = 3.27EE228 pKa = 4.34TGLADD233 pKa = 3.99ALANGAIPQPTDD245 pKa = 3.63GNIFWPVDD253 pKa = 3.29EE254 pKa = 5.23ALQLLGVSSLLDD266 pKa = 3.87HH267 pKa = 6.99YY268 pKa = 11.67DD269 pKa = 3.44SGDD272 pKa = 3.23APAIHH277 pKa = 6.51GTSGHH282 pKa = 6.55DD283 pKa = 3.68EE284 pKa = 4.49IIGTSGDD291 pKa = 3.56DD292 pKa = 3.52VIYY295 pKa = 10.83GLGGNDD301 pKa = 3.37TLTGGTADD309 pKa = 4.18DD310 pKa = 5.23FIFGGAGDD318 pKa = 4.87DD319 pKa = 3.78VLSGNAGADD328 pKa = 3.66TLDD331 pKa = 4.33GGAGDD336 pKa = 4.25DD337 pKa = 4.0WLNGGWGWDD346 pKa = 3.54EE347 pKa = 5.17LIGGSGADD355 pKa = 2.82RR356 pKa = 11.84FYY358 pKa = 11.29HH359 pKa = 6.21HH360 pKa = 6.88GVEE363 pKa = 4.38TGFGTEE369 pKa = 4.31WIHH372 pKa = 7.32DD373 pKa = 3.96FSDD376 pKa = 3.76TEE378 pKa = 4.02GDD380 pKa = 3.65RR381 pKa = 11.84LVTGASGTAGDD392 pKa = 4.08FTVSFALAEE401 pKa = 4.26GRR403 pKa = 11.84GSGDD407 pKa = 2.8IAEE410 pKa = 4.29AFIRR414 pKa = 11.84YY415 pKa = 9.03KK416 pKa = 9.9PTGQIAWVLEE426 pKa = 4.48DD427 pKa = 3.97GAGLDD432 pKa = 4.01EE433 pKa = 5.68IRR435 pKa = 11.84IQTADD440 pKa = 3.18GGFDD444 pKa = 3.67LLAA447 pKa = 5.52
MM1 pKa = 7.6TIADD5 pKa = 4.36TMTLDD10 pKa = 3.57ALAQIAHH17 pKa = 5.05QTYY20 pKa = 6.96QQRR23 pKa = 11.84YY24 pKa = 8.17NPDD27 pKa = 2.69GSINTGSLNDD37 pKa = 4.04PVAGSNGSFYY47 pKa = 10.93TEE49 pKa = 3.28ITQIVNGPLGFQARR63 pKa = 11.84AFFNEE68 pKa = 4.09TTNEE72 pKa = 3.73LVIAFTGTEE81 pKa = 4.23GFPNDD86 pKa = 3.86PAEE89 pKa = 4.14FLPDD93 pKa = 3.95FLADD97 pKa = 4.33FSLAVAGVSPQDD109 pKa = 3.51LAAQAFIKK117 pKa = 10.52QATTLAQLEE126 pKa = 4.35AGPFGSFDD134 pKa = 3.35VTYY137 pKa = 10.39VGHH140 pKa = 6.31SLGGFLAQTASASGPEE156 pKa = 4.17GEE158 pKa = 4.17VVVFNAPGAGGFLGLPEE175 pKa = 4.13NHH177 pKa = 7.32PFPEE181 pKa = 4.55DD182 pKa = 3.09NFTYY186 pKa = 10.44VYY188 pKa = 10.67SDD190 pKa = 3.55PSDD193 pKa = 3.06WGAMGGAIHH202 pKa = 6.73SAGRR206 pKa = 11.84PLSDD210 pKa = 3.57NIYY213 pKa = 9.0YY214 pKa = 10.3VPGAEE219 pKa = 4.17GHH221 pKa = 7.15DD222 pKa = 3.11ISTPDD227 pKa = 3.27EE228 pKa = 4.34TGLADD233 pKa = 3.99ALANGAIPQPTDD245 pKa = 3.63GNIFWPVDD253 pKa = 3.29EE254 pKa = 5.23ALQLLGVSSLLDD266 pKa = 3.87HH267 pKa = 6.99YY268 pKa = 11.67DD269 pKa = 3.44SGDD272 pKa = 3.23APAIHH277 pKa = 6.51GTSGHH282 pKa = 6.55DD283 pKa = 3.68EE284 pKa = 4.49IIGTSGDD291 pKa = 3.56DD292 pKa = 3.52VIYY295 pKa = 10.83GLGGNDD301 pKa = 3.37TLTGGTADD309 pKa = 4.18DD310 pKa = 5.23FIFGGAGDD318 pKa = 4.87DD319 pKa = 3.78VLSGNAGADD328 pKa = 3.66TLDD331 pKa = 4.33GGAGDD336 pKa = 4.25DD337 pKa = 4.0WLNGGWGWDD346 pKa = 3.54EE347 pKa = 5.17LIGGSGADD355 pKa = 2.82RR356 pKa = 11.84FYY358 pKa = 11.29HH359 pKa = 6.21HH360 pKa = 6.88GVEE363 pKa = 4.38TGFGTEE369 pKa = 4.31WIHH372 pKa = 7.32DD373 pKa = 3.96FSDD376 pKa = 3.76TEE378 pKa = 4.02GDD380 pKa = 3.65RR381 pKa = 11.84LVTGASGTAGDD392 pKa = 4.08FTVSFALAEE401 pKa = 4.26GRR403 pKa = 11.84GSGDD407 pKa = 2.8IAEE410 pKa = 4.29AFIRR414 pKa = 11.84YY415 pKa = 9.03KK416 pKa = 9.9PTGQIAWVLEE426 pKa = 4.48DD427 pKa = 3.97GAGLDD432 pKa = 4.01EE433 pKa = 5.68IRR435 pKa = 11.84IQTADD440 pKa = 3.18GGFDD444 pKa = 3.67LLAA447 pKa = 5.52
Molecular weight: 46.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6X7W1|A0A1M6X7W1_9RHOB Uncharacterized protein OS=Roseovarius pacificus OX=337701 GN=SAMN05444398_101319 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1358874 |
26 |
2365 |
307.2 |
33.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.877 ± 0.048 | 0.937 ± 0.011 |
6.184 ± 0.034 | 6.015 ± 0.036 |
3.665 ± 0.021 | 8.744 ± 0.04 |
2.148 ± 0.015 | 5.23 ± 0.026 |
3.064 ± 0.028 | 10.029 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.849 ± 0.018 | 2.585 ± 0.018 |
5.057 ± 0.021 | 3.242 ± 0.021 |
6.967 ± 0.036 | 5.13 ± 0.025 |
5.463 ± 0.024 | 7.185 ± 0.029 |
1.368 ± 0.015 | 2.26 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
